Pre Gene Modal
BGIBMGA014048
Annotation
PREDICTED:_SET_and_MYND_domain-containing_protein_4_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.75 Nuclear Reliability : 1.247
Sequence
CDS
ATGAGTATAGATGCTTGTTATGAAGGTGTTATTGCCAAATTAACGGATGAGGGTAAAATATTGGAAATTTCTAAGAAACTTATTAGTTTGCAAACAAATTCTGCCAGAGTTCTATTCGTATATGAGTTAATTGAATCTTTGAAAGCTTTTCCCAAAGTATTAGAGGTGAAAAAGGATGACAATGTGTCAACTTATTACCGTAACCAAGGCAACCTATTTTTTAAAGCAACTCAAAATTATGAAGCCTTACAGTTTTATAATTTGTCTTTGATACATGCACCATTGGGCACAGAGAGTTACAGTCTTGCTTTAGCCAATAGATCAGCAGTATTATTTTCGATGAAACAATATGATGAATGTATACATGATATTGATACTGTTTTTGCTATGGAATATCCGCGTGATCTAGAAAAGAAACTAAGCAAAAGGAAATCTATGTGTGAAAATTTTATATTAAAAAATAAGGAAGTAAAAGAAGACAGCACAAAAGCTAATTTCCAAGTAGCTCAAAATTACAAGTTGTTTTTAATGACGAAATGGGCCGACATGTTGTGGCTAAAGAAGATATTCAGCAAAGAATGTTCAGAAAAGGCATGGAAAGAATATCACAGAGTTGAATGTGGTTTATTAGCCACATTGGTGCATATGGAGTTTACTAAACTAGAGTTGTTGGCTCTCAGAATAGTTATAAGGGCCCGCACAGACCATAAGAATTGGCCAGAATTGTTGGAAAAATTAGTGGAGATTGAAAAAAATTTAAATACACCTTTGCGAGGATGTGAAAATTTGGGAGATAATTGGGTGTATGATTCAAAGTATTATGCTTCAATACATACACTTGAATCAAATGTGGAGAAGAGATCTGTATCGGATATTTTTCAAAAATCCATTACTGCAGCTGTATTTCTAAAATTCCTTATTGAAAATACTGATTTTATGAATGCTGATGACGCAAAACATCACAACCGGATAAAGAACTGGGTGGCTGGCACATTATTACTTCATGGCATGACTAGCCCCACTAACATGCATGGTATAATAACTAACACAGCCAACAAGAATGGAGATTATGTTGATGAACTAAACATTGGTAGTGCACCATATCCATTTTGTAGTTTGATAAATCATTCATGTGCACCAAATGTGGTCAGATATAGTAAGTTGGGCTTTGCTGAACAGATCTTGGTTGCATTGAGACCTATTAAGAAGGGTATGCAGATTTTTGACAATTATGGATCCCATTATGCAATAGAAGGCCGTCGGAGCAGACAGGAATCTCTTAAGTTCCAATACAAATTTGTTTGCGGCTGTGAAGCCTGCATAAACAACTGGCCTACTTACTTGGAAATGAGAAGGAGTAAAAATGTACCTGTTCAAATACAAAAAAGAAAAAATAAATTGTTAGATGCAAATGCTATTAATAAACTACAAAAGGGCGATAAGGGAACAGCTTTGAAATTGTTGAAGCCTCTTTGTGAACTGATGGAGCTACTTGAGCCATATTGCCCATGTTTAGAACTTGCAGATTGTCAGGAATCTTTCAAGCAGTGCCTTGCTATATGTGAGGGAGTTGTGCCTTACGGTAAGCTAATTGAATGGAATGCAATTCCACCAAAAGTTAAGTGA
Protein
MSIDACYEGVIAKLTDEGKILEISKKLISLQTNSARVLFVYELIESLKAFPKVLEVKKDDNVSTYYRNQGNLFFKATQNYEALQFYNLSLIHAPLGTESYSLALANRSAVLFSMKQYDECIHDIDTVFAMEYPRDLEKKLSKRKSMCENFILKNKEVKEDSTKANFQVAQNYKLFLMTKWADMLWLKKIFSKECSEKAWKEYHRVECGLLATLVHMEFTKLELLALRIVIRARTDHKNWPELLEKLVEIEKNLNTPLRGCENLGDNWVYDSKYYASIHTLESNVEKRSVSDIFQKSITAAVFLKFLIENTDFMNADDAKHHNRIKNWVAGTLLLHGMTSPTNMHGIITNTANKNGDYVDELNIGSAPYPFCSLINHSCAPNVVRYSKLGFAEQILVALRPIKKGMQIFDNYGSHYAIEGRRSRQESLKFQYKFVCGCEACINNWPTYLEMRRSKNVPVQIQKRKNKLLDANAINKLQKGDKGTALKLLKPLCELMELLEPYCPCLELADCQESFKQCLAICEGVVPYGKLIEWNAIPPKVK
Summary
Uniprot
A0A0N1I8M4
A0A0L7L8A6
A0A0L7L9B5
A0A194QH29
A0A2A4K7Z8
A0A2W1BMY6
+ More
A0A2H1WIT8 A0A0M5IXX9 B4J5F8 A0A151X612 B3MH29 B4KPV8 B3NS83 B4LKU5 A0A154P1E3 A0A0J9TZB5 B4HNY0 Q28XZ1 B4GH86 Q7KMH5 A0A3B0JLA9 A0A0A1XT08 A0A139WCQ9 B4P5P3 A0A0L0C194 L0CR21 L0CP86 L0CP31 L0CQD5 L0CR15 L0CP93 A0A0K8UL29 L0CPB0 W8B7G3 L0CP19 L0CP25 L0CQR4 L0CQT0 L0CP48 L0CQE1 L0CQF8 A0A0L7RBW9 A0A1Q3FCS2 B0WHZ1 R7T4C3 V4ATR0 A0A336MLV9 E0VDY8 U4U820 A0A182Y9B6 A0A151IF32 A0A182R8T1 A0A2T7PJX0 A0A084WIT6 A0A182GZU9 A0A182IZ54 B4QC82 A0A182QZT0 A0A182PX13 Q7PZC2 A0A1B0CK72 B0WI29 A0A1J1J2D0 A0A336LV57 A0A1J1HU05 A0A182LV99
A0A2H1WIT8 A0A0M5IXX9 B4J5F8 A0A151X612 B3MH29 B4KPV8 B3NS83 B4LKU5 A0A154P1E3 A0A0J9TZB5 B4HNY0 Q28XZ1 B4GH86 Q7KMH5 A0A3B0JLA9 A0A0A1XT08 A0A139WCQ9 B4P5P3 A0A0L0C194 L0CR21 L0CP86 L0CP31 L0CQD5 L0CR15 L0CP93 A0A0K8UL29 L0CPB0 W8B7G3 L0CP19 L0CP25 L0CQR4 L0CQT0 L0CP48 L0CQE1 L0CQF8 A0A0L7RBW9 A0A1Q3FCS2 B0WHZ1 R7T4C3 V4ATR0 A0A336MLV9 E0VDY8 U4U820 A0A182Y9B6 A0A151IF32 A0A182R8T1 A0A2T7PJX0 A0A084WIT6 A0A182GZU9 A0A182IZ54 B4QC82 A0A182QZT0 A0A182PX13 Q7PZC2 A0A1B0CK72 B0WI29 A0A1J1J2D0 A0A336LV57 A0A1J1HU05 A0A182LV99
Pubmed
EMBL
KQ460366
KPJ15627.1
JTDY01002367
KOB71564.1
JTDY01002104
KOB72103.1
+ More
KQ458860 KPJ04817.1 NWSH01000061 PCG80058.1 KZ149958 PZC76419.1 ODYU01008956 SOQ52985.1 CP012524 ALC42206.1 CH916367 EDW00721.1 KQ982494 KYQ55708.1 CH902619 EDV35788.1 CH933808 EDW10235.1 CH954179 EDV56385.1 CH940648 EDW60749.1 KQ434796 KZC05759.1 CM002911 KMY93095.1 CH480816 EDW47495.1 CM000071 EAL26175.1 CH479183 EDW35856.1 AF145692 AE013599 AAD38667.1 AAF58578.1 OUUW01000001 SPP74279.1 GBXI01000232 JAD14060.1 KQ971363 KYB25712.1 CM000158 EDW90840.1 JRES01001139 KNC25224.1 KC115909 AGA18631.1 KC115900 AGA18622.1 KC115901 KC115907 AGA18623.1 AGA18629.1 KC115899 KC115903 AGA18621.1 AGA18625.1 KC115904 AGA18626.1 KC115905 AGA18627.1 GDHF01025068 GDHF01004323 JAI27246.1 JAI47991.1 KC115898 KC115910 AGA18620.1 AGA18632.1 GAMC01013587 JAB92968.1 KC115897 AGA18619.1 KC115902 AGA18624.1 KC115906 AGA18628.1 KC115911 AGA18633.1 KC115912 AGA18634.1 KC115908 AGA18630.1 KC115913 AGA18635.1 KQ414617 KOC68328.1 GFDL01009726 JAV25319.1 DS231941 EDS28073.1 AMQN01015657 KB312122 ELT87827.1 KB201324 ESO97141.1 UFQS01001435 UFQT01001435 SSX10873.1 SSX30551.1 DS235088 EEB11594.1 KB632155 ERL89202.1 KQ977808 KYM99519.1 PZQS01000003 PVD33736.1 ATLV01023946 KE525347 KFB50130.1 JXUM01022095 JXUM01022096 KQ560621 KXJ81572.1 CM000362 EDX06713.1 AXCN02001095 AAAB01008986 EAA00753.4 AJWK01015852 AJWK01015853 EDS28111.1 CVRI01000066 CRL05940.1 UFQT01000128 SSX20579.1 CVRI01000021 CRK91552.1 AXCM01004284
KQ458860 KPJ04817.1 NWSH01000061 PCG80058.1 KZ149958 PZC76419.1 ODYU01008956 SOQ52985.1 CP012524 ALC42206.1 CH916367 EDW00721.1 KQ982494 KYQ55708.1 CH902619 EDV35788.1 CH933808 EDW10235.1 CH954179 EDV56385.1 CH940648 EDW60749.1 KQ434796 KZC05759.1 CM002911 KMY93095.1 CH480816 EDW47495.1 CM000071 EAL26175.1 CH479183 EDW35856.1 AF145692 AE013599 AAD38667.1 AAF58578.1 OUUW01000001 SPP74279.1 GBXI01000232 JAD14060.1 KQ971363 KYB25712.1 CM000158 EDW90840.1 JRES01001139 KNC25224.1 KC115909 AGA18631.1 KC115900 AGA18622.1 KC115901 KC115907 AGA18623.1 AGA18629.1 KC115899 KC115903 AGA18621.1 AGA18625.1 KC115904 AGA18626.1 KC115905 AGA18627.1 GDHF01025068 GDHF01004323 JAI27246.1 JAI47991.1 KC115898 KC115910 AGA18620.1 AGA18632.1 GAMC01013587 JAB92968.1 KC115897 AGA18619.1 KC115902 AGA18624.1 KC115906 AGA18628.1 KC115911 AGA18633.1 KC115912 AGA18634.1 KC115908 AGA18630.1 KC115913 AGA18635.1 KQ414617 KOC68328.1 GFDL01009726 JAV25319.1 DS231941 EDS28073.1 AMQN01015657 KB312122 ELT87827.1 KB201324 ESO97141.1 UFQS01001435 UFQT01001435 SSX10873.1 SSX30551.1 DS235088 EEB11594.1 KB632155 ERL89202.1 KQ977808 KYM99519.1 PZQS01000003 PVD33736.1 ATLV01023946 KE525347 KFB50130.1 JXUM01022095 JXUM01022096 KQ560621 KXJ81572.1 CM000362 EDX06713.1 AXCN02001095 AAAB01008986 EAA00753.4 AJWK01015852 AJWK01015853 EDS28111.1 CVRI01000066 CRL05940.1 UFQT01000128 SSX20579.1 CVRI01000021 CRK91552.1 AXCM01004284
Proteomes
UP000053240
UP000037510
UP000053268
UP000218220
UP000092553
UP000001070
+ More
UP000075809 UP000007801 UP000009192 UP000008711 UP000008792 UP000076502 UP000001292 UP000001819 UP000008744 UP000000803 UP000268350 UP000007266 UP000002282 UP000037069 UP000053825 UP000002320 UP000014760 UP000030746 UP000009046 UP000030742 UP000076408 UP000078542 UP000075900 UP000245119 UP000030765 UP000069940 UP000249989 UP000075880 UP000000304 UP000075886 UP000075885 UP000007062 UP000092461 UP000183832 UP000075883
UP000075809 UP000007801 UP000009192 UP000008711 UP000008792 UP000076502 UP000001292 UP000001819 UP000008744 UP000000803 UP000268350 UP000007266 UP000002282 UP000037069 UP000053825 UP000002320 UP000014760 UP000030746 UP000009046 UP000030742 UP000076408 UP000078542 UP000075900 UP000245119 UP000030765 UP000069940 UP000249989 UP000075880 UP000000304 UP000075886 UP000075885 UP000007062 UP000092461 UP000183832 UP000075883
Pfam
Interpro
IPR002893
Znf_MYND
+ More
IPR011990 TPR-like_helical_dom_sf
IPR001214 SET_dom
IPR008334 5'-Nucleotdase_C
IPR006146 5'-Nucleotdase_CS
IPR036907 5'-Nucleotdase_C_sf
IPR029052 Metallo-depent_PP-like
IPR006179 5_nucleotidase/apyrase
IPR019734 TPR_repeat
IPR013026 TPR-contain_dom
IPR001163 LSM_dom_euk/arc
IPR034103 Lsm8
IPR010920 LSM_dom_sf
IPR020843 PKS_ER
IPR013154 ADH_N
IPR036282 Glutathione-S-Trfase_C_sf
IPR004046 GST_C
IPR011032 GroES-like_sf
IPR013149 ADH_C
IPR010987 Glutathione-S-Trfase_C-like
IPR036291 NAD(P)-bd_dom_sf
IPR011990 TPR-like_helical_dom_sf
IPR001214 SET_dom
IPR008334 5'-Nucleotdase_C
IPR006146 5'-Nucleotdase_CS
IPR036907 5'-Nucleotdase_C_sf
IPR029052 Metallo-depent_PP-like
IPR006179 5_nucleotidase/apyrase
IPR019734 TPR_repeat
IPR013026 TPR-contain_dom
IPR001163 LSM_dom_euk/arc
IPR034103 Lsm8
IPR010920 LSM_dom_sf
IPR020843 PKS_ER
IPR013154 ADH_N
IPR036282 Glutathione-S-Trfase_C_sf
IPR004046 GST_C
IPR011032 GroES-like_sf
IPR013149 ADH_C
IPR010987 Glutathione-S-Trfase_C-like
IPR036291 NAD(P)-bd_dom_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A0N1I8M4
A0A0L7L8A6
A0A0L7L9B5
A0A194QH29
A0A2A4K7Z8
A0A2W1BMY6
+ More
A0A2H1WIT8 A0A0M5IXX9 B4J5F8 A0A151X612 B3MH29 B4KPV8 B3NS83 B4LKU5 A0A154P1E3 A0A0J9TZB5 B4HNY0 Q28XZ1 B4GH86 Q7KMH5 A0A3B0JLA9 A0A0A1XT08 A0A139WCQ9 B4P5P3 A0A0L0C194 L0CR21 L0CP86 L0CP31 L0CQD5 L0CR15 L0CP93 A0A0K8UL29 L0CPB0 W8B7G3 L0CP19 L0CP25 L0CQR4 L0CQT0 L0CP48 L0CQE1 L0CQF8 A0A0L7RBW9 A0A1Q3FCS2 B0WHZ1 R7T4C3 V4ATR0 A0A336MLV9 E0VDY8 U4U820 A0A182Y9B6 A0A151IF32 A0A182R8T1 A0A2T7PJX0 A0A084WIT6 A0A182GZU9 A0A182IZ54 B4QC82 A0A182QZT0 A0A182PX13 Q7PZC2 A0A1B0CK72 B0WI29 A0A1J1J2D0 A0A336LV57 A0A1J1HU05 A0A182LV99
A0A2H1WIT8 A0A0M5IXX9 B4J5F8 A0A151X612 B3MH29 B4KPV8 B3NS83 B4LKU5 A0A154P1E3 A0A0J9TZB5 B4HNY0 Q28XZ1 B4GH86 Q7KMH5 A0A3B0JLA9 A0A0A1XT08 A0A139WCQ9 B4P5P3 A0A0L0C194 L0CR21 L0CP86 L0CP31 L0CQD5 L0CR15 L0CP93 A0A0K8UL29 L0CPB0 W8B7G3 L0CP19 L0CP25 L0CQR4 L0CQT0 L0CP48 L0CQE1 L0CQF8 A0A0L7RBW9 A0A1Q3FCS2 B0WHZ1 R7T4C3 V4ATR0 A0A336MLV9 E0VDY8 U4U820 A0A182Y9B6 A0A151IF32 A0A182R8T1 A0A2T7PJX0 A0A084WIT6 A0A182GZU9 A0A182IZ54 B4QC82 A0A182QZT0 A0A182PX13 Q7PZC2 A0A1B0CK72 B0WI29 A0A1J1J2D0 A0A336LV57 A0A1J1HU05 A0A182LV99
PDB
5ARG
E-value=4.45898e-06,
Score=122
Ontologies
GO
PANTHER
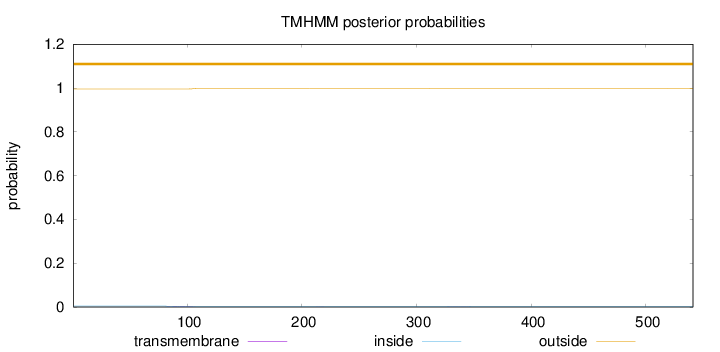
Topology
Length:
541
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06289
Exp number, first 60 AAs:
0.00067
Total prob of N-in:
0.00411
outside
1 - 541
Population Genetic Test Statistics
Pi
315.277636
Theta
187.650329
Tajima's D
0.566257
CLR
1.43243
CSRT
0.537573121343933
Interpretation
Uncertain
