Gene
KWMTBOMO01190
Pre Gene Modal
BGIBMGA007322
Annotation
PREDICTED:_uncharacterized_protein_LOC101735649_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.128
Sequence
CDS
ATGCCAGTGAACAAGTCAACACAAATTAGAGGTAACGAGATCGATAAAAGCAAACGTCGTCATAAAACTGACGACACTGTTGTTGCAACAGCGACACCAAATATCTGTGAAGATACGGTGCAATCCTCAAAAATGGAGACCCCAAACGATTACACTGGGAAACCAACAGAATTGATTCTTCAAGGCAACTCTCACTTAATGTCAAGCACCACGAGCAGTCTGGCCGATACGCCTGGTGATTCTGGTGTATTGTGTTTAGATAGTGATGCATCTGAAGCAACTAGTCAGGCCCTCATGACTCATAGTCTTGTTGGAGCAGAGGAACTAACTTGCGATAGTCTTTACGACGCTCCTCCTTCGTGCTCCCAAGACATGATAAAAAGTACCACTAGCAGCATTATGACCCGCAGTGATATTGACAATCAAAATATAGCAGCCTCAGAAGATGTTGTCCCTGAACTTGTAATTGAAACAAAAAAGTTATCACCGTTTGAAAACCTCCCTGATGTAATACTAAAAGCTTTAGAAAGTGAAAAAGGTTGTGTAGAAGAAATCAGAAAACCTGCTCTGAGATCTTGCGAACCAAAGAAATCATTATCAGAGGACATAGTATACAGACGGAGATGTAGAAAAAAGTCACAAAGTGGTCCAAGTTCACAAAAGAAAAGGGTCTCGTTTCATGAGGATATATTAAATAGCACAAAAACAGATAACATACATATTGAGCGTGGCTTCATTTCGTATAGACCAGATAGTTCATACTGTGATAGATTTAGGCAAAAGAACAATGTTGCATCTGACAGATTCTCATGGTGTGCATCTGGAGACCAAACACCAAAATATGCAGCAGATGTAGCTCAACAAACTATGTCCGATATTCTTGCATATGGAGATTCAAAAAATGATAGAAGCGGTATATTTGAATACTGTCAAAATGATGAGGCTGAAAAGCAAACTAATAAAGATAAAGACAACAATAATGAACTTAGAACAAGCGGGAAATTATATGGTTGTGATTCTAACAGCTCTGATTCTAATTTTAGTTGTGATTCAGAAACATCTTCCAGTGATTCAACTTCTTCCCATTCAAATAAAACTGAAACCTCTCCAGTGCGTAGGAAGTGTGACAAAACCCAAAAATCATACTCATGTGATGCATTTGAAAATAATGCACATGTTGCATTCATGAGGAAAACCTACTTTTCTGAAGCTGATATTGATCAATCACACAGTAGAATGAAGAAACCGCTCGAGGTTCAGCAAAGCCCAATAGCTGTAAAATCAGTTTTAAAAAAGAAACGTTACATACCTACAAATATTGTTGAAGAGAAGAAACAGAACAATAAAGTTCTAAGCTTACTGGATGCAAACAATATAATAGAGTCATTAAAAAATTTCTACAAAAACTTTCAATTCAATTTTGCTCCTGAAAGAGGTTTGCCAGAAAGTACTTTAGACTTGAACAATGTTGTTGAAGCTATTCCATGTGACATCAATCAGAACAGCAATATTTCCAAAAGCTTAGATTCTGGGTTCCATGTTGACGGGGAAGATGATTTTGTTGAAATAAATCTTAATGGTCAAACTTTGGAATCTGGTAAATCAAAAGATATTCCTGCACAGCCCAAGACTAATGTAACTGGAGTTAAGACACCTAATGGAGGATCACCTCGCCACAAACTAACACTGAATATGAAAAATCAAGATAGAAGATGTAACAAAGGAGATGTCCCAGCATCTCTTAATAAGTACATAGTTAATTGTGAAAGTACTGTGTATGAGCACAAGGGTGTATCTTATAGTTATGTACATGATACGTTCCAGACTGCATTTGATGCACCTAAAGAATCTTTTGTACCAATACCTGAGTCGACCCCAGTGAAAGAACCAACTGCAACTACAACAACAACTACAACAAACAAAGATAATTCAAAACATTCTGAAATAGATACATCAAGCAAGACATCCACACCAAAGAGGCAGTTCAATAAGAATGTCCAAGAAGAAACTGAACAAAAGAATGGTATTTCTAAACATTTGTCATCCCCTAAAAGGCAGAATGTGAACAGATATAGTCGTAAATCAGCATCAACTATCCAGAAGTTAACAGATGAAGAGAAAATTAAAGGAAATGTTGAAAACTGCTCCAACACTGACAATTCTACTTTGAAAGCCGAGAGGATGTCGCGTCTTATAGACAAAGAGGTAATAGAAAACAAAAGATTGTCTGTGAGAATGCTTACAGCAGAAGAACCATCATACTTTGACAGTTTCGAGGACAATGTCGCCATTGAATGTAACGAGAAATTAAGATTGAAAATATTTACTTATCCCAATGAAACATTGAATAGGATGATGAAAGTCCAATCACCTTATTTCATGGATGACGGATCATGGATGCCTCTATTAGTCTTCATAACAGATTACGCATTATATGTGGCCAATGTGAAGCAGAAAGGCACAGAATATGAAATCTTATGTAAATTACCCCATAATGAACTGGATGCTATAGCTGTTGGTCCCGAAGCCCAGTACATTCAGGTGATTGACATAGCAGGAAACATAGCTTGTTCGGTTGTAACGAGTGAAACTACACTCGGAGCGCGTTTAGCGTCTTCGTTGGAATGGAGCGCTCGTACATCTCCACTGCGGATACAGCGTCCGCCCGCAATGATCCCGCTCACTTGCCGGGAATTAGCCGCCGCCGTTGCCAGGAAGCGACTCGAGAAACGGGTTCCAGAAGTCCTTTTTTACGGTAGAGCGTGTTCAGAAGACGCGTGCGACAGGCTTATAGACGCACCCGGTGCCTTTGCCCCTCCACTCGAAGGATACCTCATGTGCCGCACTGACAAGAGTCGGCGCTACGAACCATGCTACTTCTTATTAAGAGCTGGGGTGCTACAGTGGGGTCCTCACAGTCCCGACGGCCTCACGCCGCTCCCGCACAGCATCGCTCTCCGCACCGCCGTGGGAGTGAGGCGGCACGTGGACGCGACCCGCAGACCGCACTGCTTCGAAGTAGCACTCGAGTCCTCTCGACTCCTACTGGCCGCACCCGATGACGCGACTGCTTCTAACTGGCTGCAGTCTCTGTTGCTACATGCCGCTCAGGCCTTAGAAGAGAAAGACGGCTACAAAAAATCGGATCGCGAGACAAGCGTACCGCCGGCAGGTTGTACGGTGCTGGTCACACCTTATGAAGTGCTCAGCGTGAGGGAGACATTAGACATGGCTTTCGTGGCCGGAGCCTCGGGGTACCTGAAGGATAGGGGGTCGGATGCTCTGCTGAAAGAATACATCGAAGAAATGCGAACCGACATCGAGATATTAGCGTGCGGATCGATAAAGAATTTGACTTCGTTTAAAATTCCGGATACAGACGAAAATTGGTGTTGCCTGGAGTGGAGCGTTTGCGAGGCGAGGGAGAGGGGCGCGGGGCTGGCGCTAGTGATGGCGAGCAGCAACGAACTCGAGCGGTTCATAGCTGCCGTCGAACAGGCATTCTGCGCACTCACCTGCGAGCCTTTCCCTCTGACCGTGGTCGAAGCCACGAAGTCATCGTGCAGCACCACACACATGAAATACGAAGCCGCCTGGTCACACATGCTCCCCTCACAGCAGTAA
Protein
MPVNKSTQIRGNEIDKSKRRHKTDDTVVATATPNICEDTVQSSKMETPNDYTGKPTELILQGNSHLMSSTTSSLADTPGDSGVLCLDSDASEATSQALMTHSLVGAEELTCDSLYDAPPSCSQDMIKSTTSSIMTRSDIDNQNIAASEDVVPELVIETKKLSPFENLPDVILKALESEKGCVEEIRKPALRSCEPKKSLSEDIVYRRRCRKKSQSGPSSQKKRVSFHEDILNSTKTDNIHIERGFISYRPDSSYCDRFRQKNNVASDRFSWCASGDQTPKYAADVAQQTMSDILAYGDSKNDRSGIFEYCQNDEAEKQTNKDKDNNNELRTSGKLYGCDSNSSDSNFSCDSETSSSDSTSSHSNKTETSPVRRKCDKTQKSYSCDAFENNAHVAFMRKTYFSEADIDQSHSRMKKPLEVQQSPIAVKSVLKKKRYIPTNIVEEKKQNNKVLSLLDANNIIESLKNFYKNFQFNFAPERGLPESTLDLNNVVEAIPCDINQNSNISKSLDSGFHVDGEDDFVEINLNGQTLESGKSKDIPAQPKTNVTGVKTPNGGSPRHKLTLNMKNQDRRCNKGDVPASLNKYIVNCESTVYEHKGVSYSYVHDTFQTAFDAPKESFVPIPESTPVKEPTATTTTTTTNKDNSKHSEIDTSSKTSTPKRQFNKNVQEETEQKNGISKHLSSPKRQNVNRYSRKSASTIQKLTDEEKIKGNVENCSNTDNSTLKAERMSRLIDKEVIENKRLSVRMLTAEEPSYFDSFEDNVAIECNEKLRLKIFTYPNETLNRMMKVQSPYFMDDGSWMPLLVFITDYALYVANVKQKGTEYEILCKLPHNELDAIAVGPEAQYIQVIDIAGNIACSVVTSETTLGARLASSLEWSARTSPLRIQRPPAMIPLTCRELAAAVARKRLEKRVPEVLFYGRACSEDACDRLIDAPGAFAPPLEGYLMCRTDKSRRYEPCYFLLRAGVLQWGPHSPDGLTPLPHSIALRTAVGVRRHVDATRRPHCFEVALESSRLLLAAPDDATASNWLQSLLLHAAQALEEKDGYKKSDRETSVPPAGCTVLVTPYEVLSVRETLDMAFVAGASGYLKDRGSDALLKEYIEEMRTDIEILACGSIKNLTSFKIPDTDENWCCLEWSVCEARERGAGLALVMASSNELERFIAAVEQAFCALTCEPFPLTVVEATKSSCSTTHMKYEAAWSHMLPSQQ
Summary
Proteomes
Pfam
PF00169 PH
Gene 3D
ProteinModelPortal
Ontologies
GO
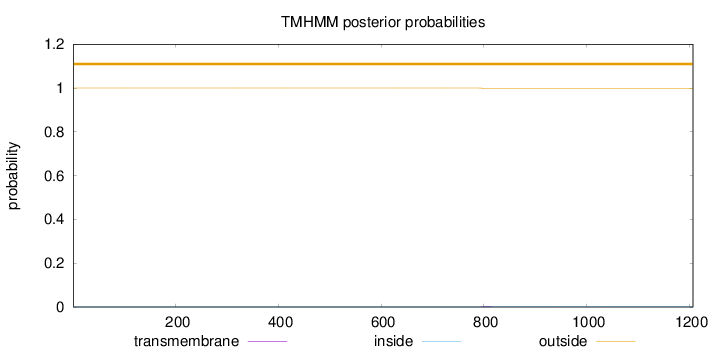
Topology
Length:
1207
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02964
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1207
Population Genetic Test Statistics
Pi
250.173135
Theta
178.821497
Tajima's D
1.112125
CLR
0.281076
CSRT
0.689065546722664
Interpretation
Uncertain
