Gene
KWMTBOMO01182 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007490
Annotation
PREDICTED:_glyceraldehyde-3-phosphate_dehydrogenase_isoform_X1_[Bombyx_mori]
Full name
Glyceraldehyde-3-phosphate dehydrogenase
Location in the cell
Cytoplasmic Reliability : 4.244
Sequence
CDS
ATGTCAAAAATTGGAATCAATGGATTTGGCCGCATTGGCCGTTTGGTGCTCCGTGCTTCTATTGAAAAGGGAGCTCAAGTGGTCGCTATAAATGACCCTTTCATCGGTCTTGACTATATGGTTTATCTTTTCAAGTATGATTCCACCCATGGCCGTTTTAAGGGCAGTGTTGAGGTTCAGGATGGATTCCTTGTTGTTAACGGTAACAAAATTGCCGTTTTCTCAGAAAGGGACCCTAAGGCCATTCCATGGGGAAAAGCTGGGGCTGAATATGTTGTAGAGTCTACTGGTGTCTTTACCACTACAGATAAAGCATCTGCTCACTTGGAGGGAGGTGCTAAAAAAGTTATTATATCAGCTCCCAGTGCTGATGCCCCCATGTTTGTTGTGGGTGTTAACCTAGAAGCTTATGACCCCTCTTTTAAGGTCATCTCAAATGCTTCTTGCACCACAAACTGTCTTGCCCCACTTGCAAAGGTTATTCATGATAACTTTGAAATTGTTGAGGGCTTGATGACTACTGTTCATGCCACAACTGCTACACAGAAAACTGTTGATGGACCTTCTGGAAAATTATGGCGTGATGGCCGTGGTGCTCAACAAAACATCATTCCTGCCTCTACTGGTGCTGCCAAAGCTGTGGGTAAGGTTATCCCTGCTCTTAATGGCAAGCTGACTGGAATGGCATTCCGCGTCCCTGTTGCTAATGTATCTGTTGTTGATCTAACTGTTCGTCTTGGAAAACCTGCAAGCTATGAAGCCATCAAGCAAAAGGTCAAGGAGGCAGCTGAAGGTCCTTTGAAGGGCATTCTCGGGTATACTGAAGATCAAGTTGTGTCCTCAGACTTCATTGGTGATTCACACTCTTCAATCTTTGATGCTGCCGCTGGAATTTCTTTGAATGACAACTTTGTGAAGCTGATCAGTTGGTACGACAATGAATATGGTTATTCCAGCAGAGTCATTGATCTCATCAAGTACATTCAATCTAAAGATTAA
Protein
MSKIGINGFGRIGRLVLRASIEKGAQVVAINDPFIGLDYMVYLFKYDSTHGRFKGSVEVQDGFLVVNGNKIAVFSERDPKAIPWGKAGAEYVVESTGVFTTTDKASAHLEGGAKKVIISAPSADAPMFVVGVNLEAYDPSFKVISNASCTTNCLAPLAKVIHDNFEIVEGLMTTVHATTATQKTVDGPSGKLWRDGRGAQQNIIPASTGAAKAVGKVIPALNGKLTGMAFRVPVANVSVVDLTVRLGKPASYEAIKQKVKEAAEGPLKGILGYTEDQVVSSDFIGDSHSSIFDAAAGISLNDNFVKLISWYDNEYGYSSRVIDLIKYIQSKD
Summary
Catalytic Activity
D-glyceraldehyde 3-phosphate + NAD(+) + phosphate = 3-phospho-D-glyceroyl phosphate + H(+) + NADH
Subunit
Homotetramer.
Similarity
Belongs to the glyceraldehyde-3-phosphate dehydrogenase family.
Feature
chain Glyceraldehyde-3-phosphate dehydrogenase
Uniprot
Q1EPM0
Q3HR37
A0A385AAY7
I4DM65
A0A0N1IPH1
A0A385AAZ4
+ More
I4DIT9 A0A385AB04 A0A1W5LB28 A0A221SMS5 A0A385AAY1 A0A385AAZ6 A0A385AAZ1 A0A212EJK4 A0A385AAZ8 A0A2A4JJ10 A0A0K0KFC7 A0A3B0E5Z2 A0A2W1BMY7 A0A385AB02 A0A0B5L5W1 S4NXR5 A0A0K2CTV2 A0A385AAY9 E7D2J4 M4Q0P2 A0A2Z5SVU8 A0A2H4WW83 F5BYH8 A0A125RM77 A0A385AAZ0 Q8MPI0 A0A0A7BYG7 A0A1W5TA01 A0A291FJ95 A0A385AB01 D7RFH6 A0A1B1LYT6 A0A0N0U4U4 E9INY0 E2ABQ6 A0A195E403 Q2VEV8 Q2VEU6 G3FP80 F4WH18 A0A0J7L8Z8 E2BXU6 Q2VEV6 R4J9E2 M9V879 A0A195EW11 Q5XXS6 A0A026WZ24 A0A1D1YHK1 A0A1B6DD01 R4J9F1 A0A286NL55 A0A195CAP2 A0A1B6DRP0 A0A0A7DNR4 A0A151XI19 Q6PPI3 A0A2R3WIG9 A0A1B6MJA5 A0A2L1GU70 A0A232FB79 K7IWP0 A0A2P0XIG5 A0A1B6LSM4 A0A1B6LLG5 A0A182MLS4 A0A182Q0I6 Q4PP91 A0A059XPP2 A0A2H8TRR6 J9K9Z1 G3LSZ8 A0A182NB09 A0A182VTT5 A0A2J7Q5L9 A0A1L8E2B2 A0A182Y8D6 A0A182P826 A0A182RCD4 A0A385AB10 A0A1B0DM89 A0A0T6AWP1 A8CWA4 A0A182K9L4 A0A385AAZ3 A0A182L624 Q7Q1U8 A0A0C5QRW7 G8CV15 A0A084VNI6 U5EWU0 A0A182XCF6 A0A182VLS6 A0A182TTA1
I4DIT9 A0A385AB04 A0A1W5LB28 A0A221SMS5 A0A385AAY1 A0A385AAZ6 A0A385AAZ1 A0A212EJK4 A0A385AAZ8 A0A2A4JJ10 A0A0K0KFC7 A0A3B0E5Z2 A0A2W1BMY7 A0A385AB02 A0A0B5L5W1 S4NXR5 A0A0K2CTV2 A0A385AAY9 E7D2J4 M4Q0P2 A0A2Z5SVU8 A0A2H4WW83 F5BYH8 A0A125RM77 A0A385AAZ0 Q8MPI0 A0A0A7BYG7 A0A1W5TA01 A0A291FJ95 A0A385AB01 D7RFH6 A0A1B1LYT6 A0A0N0U4U4 E9INY0 E2ABQ6 A0A195E403 Q2VEV8 Q2VEU6 G3FP80 F4WH18 A0A0J7L8Z8 E2BXU6 Q2VEV6 R4J9E2 M9V879 A0A195EW11 Q5XXS6 A0A026WZ24 A0A1D1YHK1 A0A1B6DD01 R4J9F1 A0A286NL55 A0A195CAP2 A0A1B6DRP0 A0A0A7DNR4 A0A151XI19 Q6PPI3 A0A2R3WIG9 A0A1B6MJA5 A0A2L1GU70 A0A232FB79 K7IWP0 A0A2P0XIG5 A0A1B6LSM4 A0A1B6LLG5 A0A182MLS4 A0A182Q0I6 Q4PP91 A0A059XPP2 A0A2H8TRR6 J9K9Z1 G3LSZ8 A0A182NB09 A0A182VTT5 A0A2J7Q5L9 A0A1L8E2B2 A0A182Y8D6 A0A182P826 A0A182RCD4 A0A385AB10 A0A1B0DM89 A0A0T6AWP1 A8CWA4 A0A182K9L4 A0A385AAZ3 A0A182L624 Q7Q1U8 A0A0C5QRW7 G8CV15 A0A084VNI6 U5EWU0 A0A182XCF6 A0A182VLS6 A0A182TTA1
EC Number
1.2.1.12
1.2.1.-
1.2.1.-
Pubmed
EMBL
BABH01036311
AB262581
BAE96011.1
DQ202316
ABA43638.2
MH592947
+ More
AXL66199.1 AK402383 BAM19005.1 KQ460366 KPJ15615.1 MH592954 AXL66206.1 AK401207 KQ458860 BAM17829.1 KPJ04805.1 MH592958 AXL66210.1 KT991373 ANA75039.1 KX668532 ASN77688.1 MH592946 AXL66198.1 MH592953 AXL66205.1 MH592945 AXL66197.1 AGBW02014444 OWR41655.1 MH592957 AXL66209.1 NWSH01001219 PCG72085.1 KF513153 AII15788.1 RBVL01000188 RKO10926.1 KZ149958 PZC76432.1 MH592955 AXL66207.1 KM923789 AJG43848.1 GAIX01012052 JAA80508.1 KT218670 ALA09390.1 MH592956 AXL66208.1 HQ012003 ADV03159.2 KC262638 ODYU01010341 AGH14258.1 SOQ55375.1 LC328973 BBA78420.1 KY514089 AUD12120.1 JF417983 AEB26314.1 KT361883 AMD40298.1 MH592950 AXL66202.1 AJ489521 CAD33827.1 KC966942 AHV85216.1 KY856799 ARF19869.1 KY039974 ATG34153.1 MH592952 AXL66204.1 HM055756 ADH51733.1 KU664394 ANS59104.1 KQ435819 KOX72508.1 GL764397 EFZ17694.1 GL438334 EFN69158.1 KQ979701 KYN19594.1 DQ205077 DQ205078 DQ205079 DQ205081 DQ205082 DQ205083 DQ205084 DQ205085 DQ205086 DQ205087 DQ205088 DQ205089 ABB90008.1 ABB90009.1 ABB90010.1 ABB90012.1 ABB90013.1 ABB90014.1 ABB90015.1 ABB90016.1 ABB90017.1 ABB90018.1 ABB90019.1 ABB90020.1 DQ205090 DQ205091 ABB90021.1 ABB90022.1 JF751029 AEO51762.1 GL888148 EGI66497.1 LBMM01000207 KMR04485.1 GL451314 EFN79503.1 DQ205080 ABB90011.1 JX273239 AGJ71764.1 KC161214 AGJ76542.1 KQ981954 KYN32082.1 AY725786 AAU95199.1 KK107061 QOIP01000014 EZA61310.1 RLU15006.1 GDJX01013818 JAT54118.1 GEDC01013762 JAS23536.1 JX273237 AGJ71762.1 KX809577 ASZ69798.1 KQ978072 KYM97266.1 GEDC01009025 JAS28273.1 KM267029 AIU97795.1 KQ982112 KYQ59965.1 AY588063 AAT01075.1 MG775273 AVQ67919.1 GEBQ01003930 JAT36047.1 KY435907 AVD73294.1 NNAY01000522 OXU27872.1 KY661337 AVA17424.1 GEBQ01013368 JAT26609.1 GEBQ01015481 JAT24496.1 AXCM01002493 AXCN02000647 DQ062222 AAY63977.1 KJ612091 AIA25511.1 GFXV01005099 MBW16904.1 ABLF02010432 JN411914 AEM75021.1 KY022438 ASL69968.1 NEVH01017557 PNF23879.1 GFDF01001362 JAV12722.1 MH592959 AXL66211.1 AJVK01006907 LJIG01022729 KRT79015.1 AJWK01006337 EU124605 ABV60323.1 MH592948 AXL66200.1 AAAB01008980 EAA13849.2 KM609486 KP676380 AJQ30110.1 ALO61090.1 HQ385974 KT724283 ADQ73875.1 AMK48125.1 ATLV01014758 KE524984 KFB39530.1 GANO01001269 JAB58602.1
AXL66199.1 AK402383 BAM19005.1 KQ460366 KPJ15615.1 MH592954 AXL66206.1 AK401207 KQ458860 BAM17829.1 KPJ04805.1 MH592958 AXL66210.1 KT991373 ANA75039.1 KX668532 ASN77688.1 MH592946 AXL66198.1 MH592953 AXL66205.1 MH592945 AXL66197.1 AGBW02014444 OWR41655.1 MH592957 AXL66209.1 NWSH01001219 PCG72085.1 KF513153 AII15788.1 RBVL01000188 RKO10926.1 KZ149958 PZC76432.1 MH592955 AXL66207.1 KM923789 AJG43848.1 GAIX01012052 JAA80508.1 KT218670 ALA09390.1 MH592956 AXL66208.1 HQ012003 ADV03159.2 KC262638 ODYU01010341 AGH14258.1 SOQ55375.1 LC328973 BBA78420.1 KY514089 AUD12120.1 JF417983 AEB26314.1 KT361883 AMD40298.1 MH592950 AXL66202.1 AJ489521 CAD33827.1 KC966942 AHV85216.1 KY856799 ARF19869.1 KY039974 ATG34153.1 MH592952 AXL66204.1 HM055756 ADH51733.1 KU664394 ANS59104.1 KQ435819 KOX72508.1 GL764397 EFZ17694.1 GL438334 EFN69158.1 KQ979701 KYN19594.1 DQ205077 DQ205078 DQ205079 DQ205081 DQ205082 DQ205083 DQ205084 DQ205085 DQ205086 DQ205087 DQ205088 DQ205089 ABB90008.1 ABB90009.1 ABB90010.1 ABB90012.1 ABB90013.1 ABB90014.1 ABB90015.1 ABB90016.1 ABB90017.1 ABB90018.1 ABB90019.1 ABB90020.1 DQ205090 DQ205091 ABB90021.1 ABB90022.1 JF751029 AEO51762.1 GL888148 EGI66497.1 LBMM01000207 KMR04485.1 GL451314 EFN79503.1 DQ205080 ABB90011.1 JX273239 AGJ71764.1 KC161214 AGJ76542.1 KQ981954 KYN32082.1 AY725786 AAU95199.1 KK107061 QOIP01000014 EZA61310.1 RLU15006.1 GDJX01013818 JAT54118.1 GEDC01013762 JAS23536.1 JX273237 AGJ71762.1 KX809577 ASZ69798.1 KQ978072 KYM97266.1 GEDC01009025 JAS28273.1 KM267029 AIU97795.1 KQ982112 KYQ59965.1 AY588063 AAT01075.1 MG775273 AVQ67919.1 GEBQ01003930 JAT36047.1 KY435907 AVD73294.1 NNAY01000522 OXU27872.1 KY661337 AVA17424.1 GEBQ01013368 JAT26609.1 GEBQ01015481 JAT24496.1 AXCM01002493 AXCN02000647 DQ062222 AAY63977.1 KJ612091 AIA25511.1 GFXV01005099 MBW16904.1 ABLF02010432 JN411914 AEM75021.1 KY022438 ASL69968.1 NEVH01017557 PNF23879.1 GFDF01001362 JAV12722.1 MH592959 AXL66211.1 AJVK01006907 LJIG01022729 KRT79015.1 AJWK01006337 EU124605 ABV60323.1 MH592948 AXL66200.1 AAAB01008980 EAA13849.2 KM609486 KP676380 AJQ30110.1 ALO61090.1 HQ385974 KT724283 ADQ73875.1 AMK48125.1 ATLV01014758 KE524984 KFB39530.1 GANO01001269 JAB58602.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000276569
+ More
UP000053105 UP000000311 UP000078492 UP000007755 UP000036403 UP000008237 UP000078541 UP000053097 UP000279307 UP000078542 UP000075809 UP000215335 UP000002358 UP000075883 UP000075886 UP000007819 UP000075884 UP000075920 UP000235965 UP000076408 UP000075885 UP000075900 UP000092462 UP000092461 UP000075881 UP000075882 UP000007062 UP000030765 UP000076407 UP000075903 UP000075902
UP000053105 UP000000311 UP000078492 UP000007755 UP000036403 UP000008237 UP000078541 UP000053097 UP000279307 UP000078542 UP000075809 UP000215335 UP000002358 UP000075883 UP000075886 UP000007819 UP000075884 UP000075920 UP000235965 UP000076408 UP000075885 UP000075900 UP000092462 UP000092461 UP000075881 UP000075882 UP000007062 UP000030765 UP000076407 UP000075903 UP000075902
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
Q1EPM0
Q3HR37
A0A385AAY7
I4DM65
A0A0N1IPH1
A0A385AAZ4
+ More
I4DIT9 A0A385AB04 A0A1W5LB28 A0A221SMS5 A0A385AAY1 A0A385AAZ6 A0A385AAZ1 A0A212EJK4 A0A385AAZ8 A0A2A4JJ10 A0A0K0KFC7 A0A3B0E5Z2 A0A2W1BMY7 A0A385AB02 A0A0B5L5W1 S4NXR5 A0A0K2CTV2 A0A385AAY9 E7D2J4 M4Q0P2 A0A2Z5SVU8 A0A2H4WW83 F5BYH8 A0A125RM77 A0A385AAZ0 Q8MPI0 A0A0A7BYG7 A0A1W5TA01 A0A291FJ95 A0A385AB01 D7RFH6 A0A1B1LYT6 A0A0N0U4U4 E9INY0 E2ABQ6 A0A195E403 Q2VEV8 Q2VEU6 G3FP80 F4WH18 A0A0J7L8Z8 E2BXU6 Q2VEV6 R4J9E2 M9V879 A0A195EW11 Q5XXS6 A0A026WZ24 A0A1D1YHK1 A0A1B6DD01 R4J9F1 A0A286NL55 A0A195CAP2 A0A1B6DRP0 A0A0A7DNR4 A0A151XI19 Q6PPI3 A0A2R3WIG9 A0A1B6MJA5 A0A2L1GU70 A0A232FB79 K7IWP0 A0A2P0XIG5 A0A1B6LSM4 A0A1B6LLG5 A0A182MLS4 A0A182Q0I6 Q4PP91 A0A059XPP2 A0A2H8TRR6 J9K9Z1 G3LSZ8 A0A182NB09 A0A182VTT5 A0A2J7Q5L9 A0A1L8E2B2 A0A182Y8D6 A0A182P826 A0A182RCD4 A0A385AB10 A0A1B0DM89 A0A0T6AWP1 A8CWA4 A0A182K9L4 A0A385AAZ3 A0A182L624 Q7Q1U8 A0A0C5QRW7 G8CV15 A0A084VNI6 U5EWU0 A0A182XCF6 A0A182VLS6 A0A182TTA1
I4DIT9 A0A385AB04 A0A1W5LB28 A0A221SMS5 A0A385AAY1 A0A385AAZ6 A0A385AAZ1 A0A212EJK4 A0A385AAZ8 A0A2A4JJ10 A0A0K0KFC7 A0A3B0E5Z2 A0A2W1BMY7 A0A385AB02 A0A0B5L5W1 S4NXR5 A0A0K2CTV2 A0A385AAY9 E7D2J4 M4Q0P2 A0A2Z5SVU8 A0A2H4WW83 F5BYH8 A0A125RM77 A0A385AAZ0 Q8MPI0 A0A0A7BYG7 A0A1W5TA01 A0A291FJ95 A0A385AB01 D7RFH6 A0A1B1LYT6 A0A0N0U4U4 E9INY0 E2ABQ6 A0A195E403 Q2VEV8 Q2VEU6 G3FP80 F4WH18 A0A0J7L8Z8 E2BXU6 Q2VEV6 R4J9E2 M9V879 A0A195EW11 Q5XXS6 A0A026WZ24 A0A1D1YHK1 A0A1B6DD01 R4J9F1 A0A286NL55 A0A195CAP2 A0A1B6DRP0 A0A0A7DNR4 A0A151XI19 Q6PPI3 A0A2R3WIG9 A0A1B6MJA5 A0A2L1GU70 A0A232FB79 K7IWP0 A0A2P0XIG5 A0A1B6LSM4 A0A1B6LLG5 A0A182MLS4 A0A182Q0I6 Q4PP91 A0A059XPP2 A0A2H8TRR6 J9K9Z1 G3LSZ8 A0A182NB09 A0A182VTT5 A0A2J7Q5L9 A0A1L8E2B2 A0A182Y8D6 A0A182P826 A0A182RCD4 A0A385AB10 A0A1B0DM89 A0A0T6AWP1 A8CWA4 A0A182K9L4 A0A385AAZ3 A0A182L624 Q7Q1U8 A0A0C5QRW7 G8CV15 A0A084VNI6 U5EWU0 A0A182XCF6 A0A182VLS6 A0A182TTA1
PDB
4WNC
E-value=1.20982e-139,
Score=1271
Ontologies
GO
PANTHER
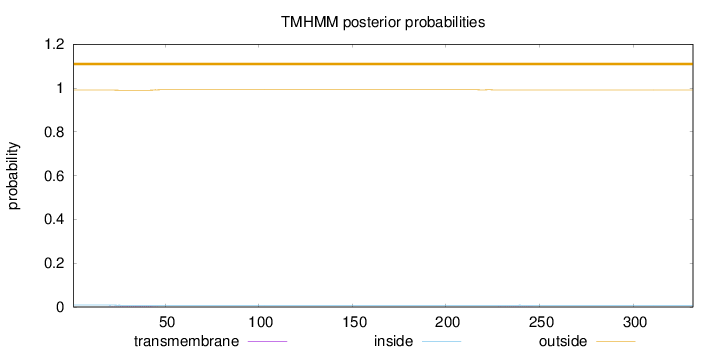
Topology
Length:
332
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0454900000000001
Exp number, first 60 AAs:
0.03973
Total prob of N-in:
0.00919
outside
1 - 332
Population Genetic Test Statistics
Pi
121.826189
Theta
213.514595
Tajima's D
-1.260812
CLR
0.359942
CSRT
0.095845207739613
Interpretation
Uncertain
