Pre Gene Modal
BGIBMGA007489
Annotation
PREDICTED:_PI-PLC_X_domain-containing_protein_3_isoform_X2_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.002 PlasmaMembrane Reliability : 1.043
Sequence
CDS
ATGAGTGAGGATGGAGCGGCGCAAAACTTTCGTCTTGAATCATGGATGAGTGATCTACCAATACAGCTGAAGGATATACCAATAATCTACTTAGCCATACCTGGCAGTCATGACTCAATGACCTACGGCATCACAAAATCTAGTGGCCTAGCACCTGACGCTGAACCTATTCTTCATCGCTTGTACCCCTTATTCCGAGGTACGATCTTACGATGGACAATAACACAAGCTCTTGACACCAAGCAACAACTACTTATTGGCATTAGATATTTAGACCTCAGGATTGCTACAAAATCTGGTGAAGACCATTTTTATTTCACACATGGTGTTTACGCTGCTGAAATAACAGAGGCACTGAAGCAAGTCAAAGAGTTCGTTGATGAACATCCTGGAGAGGTAGTGATACTTGACTGCCAACATTTCTATGGCTTCCAACCTGAAGACCACCAGAGACTGATGAGGCTCCTCCTCAATATGTATGGTCCCATATTGGTACCACGGCAGCTTGATTTGAAAGCTGTTACTCTGAATGCTCTACAACGACTAAAGCAGCAAGTGATTGTGGTGTACCGCCACGAGTCTGTGTACGCGACGGGCGAGTTCTGGCCGCCGCAGACGTTGCCGTCGCCGTGGCCGCGCCAGGACCGCGCGCCGGGGCTGCTCGCGTTCCTGGGCGCGCTGTGTCGGCACCCGAGCGCCGGCTTCGTGCACCAGGCCGTGCTCACGCCCACCCCGGCCTTCATCGTGCTCAGATGGATATCAAGTCTCCGCGATAAATGCGCATTGCCAGTCATCAACGATGTGCTACCAAAATTATCTGATTTCTCTCCCGGACCCCCGGCACCGGACAACGAGACGTTCTCGCGGGCGCCCGTCAACGTAGTCATCGCAGACTTCGTTGACATCAATGACTCGATATTCTCAAAAACTATAATACAGTTGAACATGAAGTTGTTGAGAAACACAGATGTCCTTTATCACACCAACGAATAG
Protein
MSEDGAAQNFRLESWMSDLPIQLKDIPIIYLAIPGSHDSMTYGITKSSGLAPDAEPILHRLYPLFRGTILRWTITQALDTKQQLLIGIRYLDLRIATKSGEDHFYFTHGVYAAEITEALKQVKEFVDEHPGEVVILDCQHFYGFQPEDHQRLMRLLLNMYGPILVPRQLDLKAVTLNALQRLKQQVIVVYRHESVYATGEFWPPQTLPSPWPRQDRAPGLLAFLGALCRHPSAGFVHQAVLTPTPAFIVLRWISSLRDKCALPVINDVLPKLSDFSPGPPAPDNETFSRAPVNVVIADFVDINDSIFSKTIIQLNMKLLRNTDVLYHTNE
Summary
Uniprot
H9JD92
A0A2W1BS13
A0A2A4JJD3
A0A2A4JKZ8
A0A1E1WN44
A0A1E1W2H5
+ More
A0A194QGY4 A0A0N1I8M0 A0A2H1WST6 A0A212EJJ1 D6WW38 A0A1S4FWX6 A0A182GKD8 A0A1B6F4D0 A0A1B6J506 A0A1B0EZZ8 Q16LR9 B0WPF7 A0A182W8P7 A0A182LSJ7 A0A182RWP4 A0A084VSN8 A0A182NP06 A0A182S5U1 A0A1B6LKK9 A0A182Y682 A0A2J7QZV8 A0A1Q3F0R8 A0A1B0DD55 A0A182Q186 A0A067RE23 A0A182VIK2 A0A182X2F9 A0A182KZ65 Q7Q123 A0A182HR74 A0A0V0G8L7 A0A182F359 A0A182TZI5 A0A182PVR0 W5JF14 A0A182IYF0 A0A1Y1L8T1 A0A1W4VWT9 A0A1J1II62 A0A069DYL1 D6WW39 A0A3B0KHU3 B4GT66 N6T8D1 Q29KK9 U4UIM9 A0A0J9R5W6 B4PBH8 B3NL03 A0A0N7Z8P3 A0A1B6DUS4 B4QAH2 B4I634 Q8SXL1 A0A0K8TKI5 B4M9M1 Q9VIP3 B3ML25 A0A336M9J6 A0A0M4EQ84 B4MTM2 A0A158P2H7 A0A3Q0J2Y0 A0A2R7WV81 B4KEV7 A0A151XAW5 A0A224Z513 L7M120 A0A2P8XLT1 A0A232EUV3 A0A195DHZ1 E2BJV0 A0A1E1XDV0 B4JQ93 A0A195BST0 A0A034VV58 E9J3X8 A0A131YUH2 B7QN53 A0A2P8YQG1 A0A131XXL2 A0A195CLM3 V5HAP6 A0A147BH03 A0A026WF91 A0A1Z5L633 A0A293L9X4 A0A0C9R848 A0A0K8U8P9 A0A2P6L846 A0A0M8ZZ38 A0A1A9XP03 A0A088AEK8
A0A194QGY4 A0A0N1I8M0 A0A2H1WST6 A0A212EJJ1 D6WW38 A0A1S4FWX6 A0A182GKD8 A0A1B6F4D0 A0A1B6J506 A0A1B0EZZ8 Q16LR9 B0WPF7 A0A182W8P7 A0A182LSJ7 A0A182RWP4 A0A084VSN8 A0A182NP06 A0A182S5U1 A0A1B6LKK9 A0A182Y682 A0A2J7QZV8 A0A1Q3F0R8 A0A1B0DD55 A0A182Q186 A0A067RE23 A0A182VIK2 A0A182X2F9 A0A182KZ65 Q7Q123 A0A182HR74 A0A0V0G8L7 A0A182F359 A0A182TZI5 A0A182PVR0 W5JF14 A0A182IYF0 A0A1Y1L8T1 A0A1W4VWT9 A0A1J1II62 A0A069DYL1 D6WW39 A0A3B0KHU3 B4GT66 N6T8D1 Q29KK9 U4UIM9 A0A0J9R5W6 B4PBH8 B3NL03 A0A0N7Z8P3 A0A1B6DUS4 B4QAH2 B4I634 Q8SXL1 A0A0K8TKI5 B4M9M1 Q9VIP3 B3ML25 A0A336M9J6 A0A0M4EQ84 B4MTM2 A0A158P2H7 A0A3Q0J2Y0 A0A2R7WV81 B4KEV7 A0A151XAW5 A0A224Z513 L7M120 A0A2P8XLT1 A0A232EUV3 A0A195DHZ1 E2BJV0 A0A1E1XDV0 B4JQ93 A0A195BST0 A0A034VV58 E9J3X8 A0A131YUH2 B7QN53 A0A2P8YQG1 A0A131XXL2 A0A195CLM3 V5HAP6 A0A147BH03 A0A026WF91 A0A1Z5L633 A0A293L9X4 A0A0C9R848 A0A0K8U8P9 A0A2P6L846 A0A0M8ZZ38 A0A1A9XP03 A0A088AEK8
Pubmed
19121390
28756777
26354079
22118469
18362917
19820115
+ More
26483478 17510324 24438588 25244985 24845553 20966253 12364791 14747013 17210077 20920257 23761445 28004739 26334808 17994087 23537049 15632085 22936249 17550304 27129103 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 28797301 25576852 29403074 28648823 20798317 28503490 25348373 21282665 26830274 25765539 29652888 24508170 30249741 28528879
26483478 17510324 24438588 25244985 24845553 20966253 12364791 14747013 17210077 20920257 23761445 28004739 26334808 17994087 23537049 15632085 22936249 17550304 27129103 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 28797301 25576852 29403074 28648823 20798317 28503490 25348373 21282665 26830274 25765539 29652888 24508170 30249741 28528879
EMBL
BABH01036311
KZ149958
PZC76434.1
NWSH01001219
PCG72081.1
PCG72080.1
+ More
GDQN01002707 JAT88347.1 GDQN01009866 JAT81188.1 KQ458860 KPJ04803.1 KQ460366 KPJ15613.1 ODYU01010796 SOQ56121.1 AGBW02014444 OWR41657.1 KQ971361 EFA08659.1 JXUM01069814 KQ562571 KXJ75575.1 GECZ01024732 JAS45037.1 GECU01013454 JAS94252.1 AJWK01030842 CH477894 EAT35279.1 DS232023 EDS32336.1 AXCM01000351 ATLV01016119 KE525057 KFB40982.1 GEBQ01015734 JAT24243.1 NEVH01009067 PNF34116.1 GFDL01013895 JAV21150.1 AJVK01031645 AXCN02001960 KK852521 KDR22106.1 AAAB01008980 EAA13925.4 APCN01001654 GECL01002016 JAP04108.1 ADMH02001597 ETN61918.1 GEZM01066301 GEZM01066300 GEZM01066299 JAV67986.1 CVRI01000054 CRK99941.1 GBGD01002200 JAC86689.1 EFA08660.1 OUUW01000010 SPP85969.1 CH479189 EDW25575.1 APGK01039948 KB740975 ENN76489.1 CH379061 EAL33165.2 KB632186 ERL89775.1 CM002910 KMY91129.1 CM000158 EDW90492.1 CH954179 EDV54581.1 GDKW01002787 JAI53808.1 GEDC01007862 JAS29436.1 CM000361 EDX05589.1 CH480822 EDW55840.1 AY089577 AAL90315.1 GDAI01002789 JAI14814.1 CH940654 EDW57897.1 AE014134 BT044106 AAF53873.2 ACH92171.1 AGB93153.1 CH902620 EDV31643.1 UFQT01000758 SSX27012.1 CP012523 ALC38882.1 CH963852 EDW75461.1 ADTU01006753 KK855666 PTY23462.1 CH933807 EDW11926.1 KQ982335 KYQ57479.1 GFPF01010557 MAA21703.1 GACK01007013 JAA58021.1 PYGN01001761 PSN32955.1 NNAY01002087 OXU22116.1 KQ980824 KYN12457.1 GL448661 EFN84033.1 GFAC01001819 JAT97369.1 CH916372 EDV99073.1 KQ976419 KYM89337.1 GAKP01012623 JAC46329.1 GL768069 EFZ12419.1 GEDV01005703 JAP82854.1 ABJB011041770 DS976400 EEC20275.1 PYGN01000432 PSN46490.1 GEFM01005485 JAP70311.1 KQ977595 KYN01600.1 GANP01010149 JAB74319.1 GEGO01005366 JAR90038.1 KK107274 QOIP01000008 EZA53679.1 RLU19211.1 GFJQ02004312 JAW02658.1 GFWV01000384 MAA25114.1 GBYB01004210 JAG73977.1 GDHF01029280 JAI23034.1 MWRG01001093 PRD34749.1 KQ435817 KOX72553.1
GDQN01002707 JAT88347.1 GDQN01009866 JAT81188.1 KQ458860 KPJ04803.1 KQ460366 KPJ15613.1 ODYU01010796 SOQ56121.1 AGBW02014444 OWR41657.1 KQ971361 EFA08659.1 JXUM01069814 KQ562571 KXJ75575.1 GECZ01024732 JAS45037.1 GECU01013454 JAS94252.1 AJWK01030842 CH477894 EAT35279.1 DS232023 EDS32336.1 AXCM01000351 ATLV01016119 KE525057 KFB40982.1 GEBQ01015734 JAT24243.1 NEVH01009067 PNF34116.1 GFDL01013895 JAV21150.1 AJVK01031645 AXCN02001960 KK852521 KDR22106.1 AAAB01008980 EAA13925.4 APCN01001654 GECL01002016 JAP04108.1 ADMH02001597 ETN61918.1 GEZM01066301 GEZM01066300 GEZM01066299 JAV67986.1 CVRI01000054 CRK99941.1 GBGD01002200 JAC86689.1 EFA08660.1 OUUW01000010 SPP85969.1 CH479189 EDW25575.1 APGK01039948 KB740975 ENN76489.1 CH379061 EAL33165.2 KB632186 ERL89775.1 CM002910 KMY91129.1 CM000158 EDW90492.1 CH954179 EDV54581.1 GDKW01002787 JAI53808.1 GEDC01007862 JAS29436.1 CM000361 EDX05589.1 CH480822 EDW55840.1 AY089577 AAL90315.1 GDAI01002789 JAI14814.1 CH940654 EDW57897.1 AE014134 BT044106 AAF53873.2 ACH92171.1 AGB93153.1 CH902620 EDV31643.1 UFQT01000758 SSX27012.1 CP012523 ALC38882.1 CH963852 EDW75461.1 ADTU01006753 KK855666 PTY23462.1 CH933807 EDW11926.1 KQ982335 KYQ57479.1 GFPF01010557 MAA21703.1 GACK01007013 JAA58021.1 PYGN01001761 PSN32955.1 NNAY01002087 OXU22116.1 KQ980824 KYN12457.1 GL448661 EFN84033.1 GFAC01001819 JAT97369.1 CH916372 EDV99073.1 KQ976419 KYM89337.1 GAKP01012623 JAC46329.1 GL768069 EFZ12419.1 GEDV01005703 JAP82854.1 ABJB011041770 DS976400 EEC20275.1 PYGN01000432 PSN46490.1 GEFM01005485 JAP70311.1 KQ977595 KYN01600.1 GANP01010149 JAB74319.1 GEGO01005366 JAR90038.1 KK107274 QOIP01000008 EZA53679.1 RLU19211.1 GFJQ02004312 JAW02658.1 GFWV01000384 MAA25114.1 GBYB01004210 JAG73977.1 GDHF01029280 JAI23034.1 MWRG01001093 PRD34749.1 KQ435817 KOX72553.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000007266
+ More
UP000069940 UP000249989 UP000092461 UP000008820 UP000002320 UP000075920 UP000075883 UP000075900 UP000030765 UP000075884 UP000075901 UP000076408 UP000235965 UP000092462 UP000075886 UP000027135 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000069272 UP000075902 UP000075885 UP000000673 UP000075880 UP000192221 UP000183832 UP000268350 UP000008744 UP000019118 UP000001819 UP000030742 UP000002282 UP000008711 UP000000304 UP000001292 UP000008792 UP000000803 UP000007801 UP000092553 UP000007798 UP000005205 UP000079169 UP000009192 UP000075809 UP000245037 UP000215335 UP000078492 UP000008237 UP000001070 UP000078540 UP000001555 UP000078542 UP000053097 UP000279307 UP000053105 UP000092443 UP000005203
UP000069940 UP000249989 UP000092461 UP000008820 UP000002320 UP000075920 UP000075883 UP000075900 UP000030765 UP000075884 UP000075901 UP000076408 UP000235965 UP000092462 UP000075886 UP000027135 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000069272 UP000075902 UP000075885 UP000000673 UP000075880 UP000192221 UP000183832 UP000268350 UP000008744 UP000019118 UP000001819 UP000030742 UP000002282 UP000008711 UP000000304 UP000001292 UP000008792 UP000000803 UP000007801 UP000092553 UP000007798 UP000005205 UP000079169 UP000009192 UP000075809 UP000245037 UP000215335 UP000078492 UP000008237 UP000001070 UP000078540 UP000001555 UP000078542 UP000053097 UP000279307 UP000053105 UP000092443 UP000005203
Interpro
SUPFAM
SSF51695
SSF51695
Gene 3D
ProteinModelPortal
H9JD92
A0A2W1BS13
A0A2A4JJD3
A0A2A4JKZ8
A0A1E1WN44
A0A1E1W2H5
+ More
A0A194QGY4 A0A0N1I8M0 A0A2H1WST6 A0A212EJJ1 D6WW38 A0A1S4FWX6 A0A182GKD8 A0A1B6F4D0 A0A1B6J506 A0A1B0EZZ8 Q16LR9 B0WPF7 A0A182W8P7 A0A182LSJ7 A0A182RWP4 A0A084VSN8 A0A182NP06 A0A182S5U1 A0A1B6LKK9 A0A182Y682 A0A2J7QZV8 A0A1Q3F0R8 A0A1B0DD55 A0A182Q186 A0A067RE23 A0A182VIK2 A0A182X2F9 A0A182KZ65 Q7Q123 A0A182HR74 A0A0V0G8L7 A0A182F359 A0A182TZI5 A0A182PVR0 W5JF14 A0A182IYF0 A0A1Y1L8T1 A0A1W4VWT9 A0A1J1II62 A0A069DYL1 D6WW39 A0A3B0KHU3 B4GT66 N6T8D1 Q29KK9 U4UIM9 A0A0J9R5W6 B4PBH8 B3NL03 A0A0N7Z8P3 A0A1B6DUS4 B4QAH2 B4I634 Q8SXL1 A0A0K8TKI5 B4M9M1 Q9VIP3 B3ML25 A0A336M9J6 A0A0M4EQ84 B4MTM2 A0A158P2H7 A0A3Q0J2Y0 A0A2R7WV81 B4KEV7 A0A151XAW5 A0A224Z513 L7M120 A0A2P8XLT1 A0A232EUV3 A0A195DHZ1 E2BJV0 A0A1E1XDV0 B4JQ93 A0A195BST0 A0A034VV58 E9J3X8 A0A131YUH2 B7QN53 A0A2P8YQG1 A0A131XXL2 A0A195CLM3 V5HAP6 A0A147BH03 A0A026WF91 A0A1Z5L633 A0A293L9X4 A0A0C9R848 A0A0K8U8P9 A0A2P6L846 A0A0M8ZZ38 A0A1A9XP03 A0A088AEK8
A0A194QGY4 A0A0N1I8M0 A0A2H1WST6 A0A212EJJ1 D6WW38 A0A1S4FWX6 A0A182GKD8 A0A1B6F4D0 A0A1B6J506 A0A1B0EZZ8 Q16LR9 B0WPF7 A0A182W8P7 A0A182LSJ7 A0A182RWP4 A0A084VSN8 A0A182NP06 A0A182S5U1 A0A1B6LKK9 A0A182Y682 A0A2J7QZV8 A0A1Q3F0R8 A0A1B0DD55 A0A182Q186 A0A067RE23 A0A182VIK2 A0A182X2F9 A0A182KZ65 Q7Q123 A0A182HR74 A0A0V0G8L7 A0A182F359 A0A182TZI5 A0A182PVR0 W5JF14 A0A182IYF0 A0A1Y1L8T1 A0A1W4VWT9 A0A1J1II62 A0A069DYL1 D6WW39 A0A3B0KHU3 B4GT66 N6T8D1 Q29KK9 U4UIM9 A0A0J9R5W6 B4PBH8 B3NL03 A0A0N7Z8P3 A0A1B6DUS4 B4QAH2 B4I634 Q8SXL1 A0A0K8TKI5 B4M9M1 Q9VIP3 B3ML25 A0A336M9J6 A0A0M4EQ84 B4MTM2 A0A158P2H7 A0A3Q0J2Y0 A0A2R7WV81 B4KEV7 A0A151XAW5 A0A224Z513 L7M120 A0A2P8XLT1 A0A232EUV3 A0A195DHZ1 E2BJV0 A0A1E1XDV0 B4JQ93 A0A195BST0 A0A034VV58 E9J3X8 A0A131YUH2 B7QN53 A0A2P8YQG1 A0A131XXL2 A0A195CLM3 V5HAP6 A0A147BH03 A0A026WF91 A0A1Z5L633 A0A293L9X4 A0A0C9R848 A0A0K8U8P9 A0A2P6L846 A0A0M8ZZ38 A0A1A9XP03 A0A088AEK8
PDB
1AOD
E-value=2.96852e-05,
Score=112
Ontologies
GO
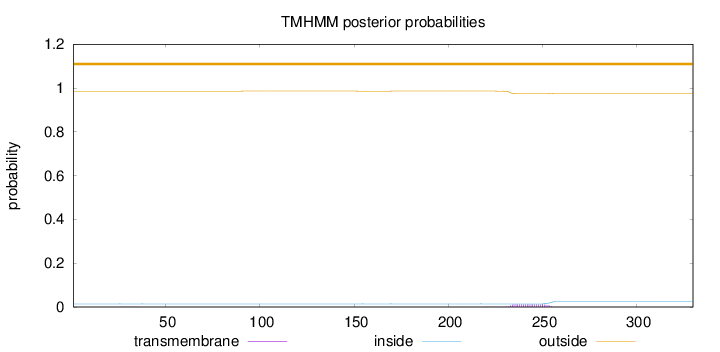
Topology
Length:
330
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2699
Exp number, first 60 AAs:
0.00356
Total prob of N-in:
0.01412
outside
1 - 330
Population Genetic Test Statistics
Pi
201.217525
Theta
153.195191
Tajima's D
0.630273
CLR
0.276014
CSRT
0.55807209639518
Interpretation
Uncertain
