Gene
KWMTBOMO01175
Pre Gene Modal
BGIBMGA007449
Annotation
PREDICTED:_NAD_kinase-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.072 PlasmaMembrane Reliability : 1.084
Sequence
CDS
ATGGTCATGCAAATCCAGGATCCGGCGTCGCAGAGGCTGACCTGGTACAAGCCTCCCCTCACCGTGCTGGTCATCAAGAAGGTTCACGACGCCCAGATACTGGCACCCTTCGTTCAGCTAGTACACTGGCTCGTTCATGACAAAAGCATGGTGGTGTTCGTCGAGGCGGCAGTTTTAGACGACACACTCCTTGCTGAGTATGGAGACTTTACGTCGGTGAAGGAACGTCTGATGACTTTCAGAGCTAGCACCGATGATCTCACTGATAAGATCGACTTCATCATATGCCTGGGGGGTGATGGAACTCTGTTGCATGCTAGTTCACTTTTCCAGCAATCAGTGCCGCCGGTGATGGCGTTCCACTTGGGCTCGTTGGGTTTTCTGACGCCCTTCGAGTTCAACAACTTTCAGGAGCAAGTCATGAACGTTTTAGAAGGACACGCGGCTCTAACTTTAAGGTCCCGTTTGCAATGCGTAGTGCTTCGAAAGAGTCAGGATGACAATAAAGACAAGAAGAAGCCGACCACGATACTGGTGCTGAACGAGGTTGTCGTGGACCGAGGACCATCGCCGTATTTATCCAATATAGATCTGTTCCTCGACGGGAAACACATCACGTCTGTGCAAGGAGACGGGCTGATAGTGTCGACGCCGACGGGCAGCACGGCGTACGCGGTGGCGGCGGGCGCGAGCATGATCCACCCGTCGGTGCCCGCCATCATGGTCACGCCAATCTGCCCGCATTCGTTGTCCTTCAGACCCATCGTAGTGCCGGCCGGCGTCGAGCTCAAGATAATGCCTAGCCCCGAGGCCCGTAATTCAGCCAGCGTGTCGTTCGACGGCCGCTCCCAACTCACTCTCAGACCCGGCGATAGTCTCTACGTGACCACGTCCGTGTATCCGGTGCCGTCGATATGCGCCCAGGATCAGATCTCGGATTGGTTCGACTCGCTCGCCGAGTGCCTCCATTGGAACGTTAGGAAGAAACAGAAGCAGATGGACGAGCTGAGCGACCTCACGCACTCGTCGAGCGACAACCTCGAGGAGCTAGACAATAACCACGATGAAAGACCATCAGATACGTGA
Protein
MVMQIQDPASQRLTWYKPPLTVLVIKKVHDAQILAPFVQLVHWLVHDKSMVVFVEAAVLDDTLLAEYGDFTSVKERLMTFRASTDDLTDKIDFIICLGGDGTLLHASSLFQQSVPPVMAFHLGSLGFLTPFEFNNFQEQVMNVLEGHAALTLRSRLQCVVLRKSQDDNKDKKKPTTILVLNEVVVDRGPSPYLSNIDLFLDGKHITSVQGDGLIVSTPTGSTAYAVAAGASMIHPSVPAIMVTPICPHSLSFRPIVVPAGVELKIMPSPEARNSASVSFDGRSQLTLRPGDSLYVTTSVYPVPSICAQDQISDWFDSLAECLHWNVRKKQKQMDELSDLTHSSSDNLEELDNNHDERPSDT
Summary
Uniprot
H9JD52
W8FNS9
W8FPH0
I4DMW8
A0A0N1PJB8
A0A194QFZ6
+ More
A0A2H1WUU3 A0A2W1BTA1 A0A212EJL7 V5G4S7 A0A1B6CXK4 A0A2J7QZQ3 A0A2J7QZR9 A0A2J7QZQ2 A0A2A4J3B5 A0A1B6E8I6 A0A069DT24 A0A2J7QZQ5 A0A2J7QZT2 A0A1B6MH08 A0A224XE60 A0A0V0G7T0 A0A1S3CYN3 A0A1B6LHG2 A0A1B6L3K8 A0A2J7QZS9 A0A1B6MIM4 A0A1B6DV81 A0A1S3CZZ4 A0A1S3CYP3 A0A1S3CYN8 A0A1S3CYM6 V5GLU2 A0A1S3CYM1 A0A1S3CYT3 A0A3Q0IQ79 A0A1S3CYQ9 A0A2J7QZQ7 A0A3Q0IQ14 A0A1S3CYL6 A0A1S3CYQ4 A0A1W4WVS0 A0A1S3CYS8 E9GAJ9 A0A232F5G1 A0A0P5G0Z2 A0A0P5HPC8 E0VE17 A0A1Y1LES9 A0A154P1H9 A0A0P5GI57 A0A0M9AAZ6 A0A310SKZ1 A0A0N8CU87 A0A0P4ZG78 A0A1W4X6L1 A0A0A9XZS4 D6WXV3 N6TPP3 A0A336LT55 A0A0A9Y5D0 K7ING6 A0A1Y1LD81 A0A2A4J3M7 A0A146LT56 A0A0A9XZR9 A0A0P6DZX2 A0A0P5JJL6 A0A0P5CEN9 E2A820 A0A0L7LQR6 A0A0P6HD38 A0A336KA94 A0A0P5LUN3 A0A088AM99 A0A158NST4 A0A034VML6 A0A0P5S5X8 A0A034VHK1 A0A1J1I5R3 A0A1J1I5K4 A0A336LWI5 A0A087U528 A0A034VJX2 A0A151I062 A0A158NST3 A0A0A1WSZ1 F4WSH5 A0A034VLW0 A0A0K8UWE6 A0A151X623 A0A195DPF4 E9IQ04 A0A151IF27 A0A2L2Y5B2 A0A2L2Y5E5 T1IL69 A0A1L8E1S5 A0A1B0GQR1 A0A1B0GKN9
A0A2H1WUU3 A0A2W1BTA1 A0A212EJL7 V5G4S7 A0A1B6CXK4 A0A2J7QZQ3 A0A2J7QZR9 A0A2J7QZQ2 A0A2A4J3B5 A0A1B6E8I6 A0A069DT24 A0A2J7QZQ5 A0A2J7QZT2 A0A1B6MH08 A0A224XE60 A0A0V0G7T0 A0A1S3CYN3 A0A1B6LHG2 A0A1B6L3K8 A0A2J7QZS9 A0A1B6MIM4 A0A1B6DV81 A0A1S3CZZ4 A0A1S3CYP3 A0A1S3CYN8 A0A1S3CYM6 V5GLU2 A0A1S3CYM1 A0A1S3CYT3 A0A3Q0IQ79 A0A1S3CYQ9 A0A2J7QZQ7 A0A3Q0IQ14 A0A1S3CYL6 A0A1S3CYQ4 A0A1W4WVS0 A0A1S3CYS8 E9GAJ9 A0A232F5G1 A0A0P5G0Z2 A0A0P5HPC8 E0VE17 A0A1Y1LES9 A0A154P1H9 A0A0P5GI57 A0A0M9AAZ6 A0A310SKZ1 A0A0N8CU87 A0A0P4ZG78 A0A1W4X6L1 A0A0A9XZS4 D6WXV3 N6TPP3 A0A336LT55 A0A0A9Y5D0 K7ING6 A0A1Y1LD81 A0A2A4J3M7 A0A146LT56 A0A0A9XZR9 A0A0P6DZX2 A0A0P5JJL6 A0A0P5CEN9 E2A820 A0A0L7LQR6 A0A0P6HD38 A0A336KA94 A0A0P5LUN3 A0A088AM99 A0A158NST4 A0A034VML6 A0A0P5S5X8 A0A034VHK1 A0A1J1I5R3 A0A1J1I5K4 A0A336LWI5 A0A087U528 A0A034VJX2 A0A151I062 A0A158NST3 A0A0A1WSZ1 F4WSH5 A0A034VLW0 A0A0K8UWE6 A0A151X623 A0A195DPF4 E9IQ04 A0A151IF27 A0A2L2Y5B2 A0A2L2Y5E5 T1IL69 A0A1L8E1S5 A0A1B0GQR1 A0A1B0GKN9
Pubmed
EMBL
BABH01036295
KF924769
AHK10564.1
KF924768
AHK10563.1
AK402636
+ More
BAM19258.1 KQ460366 KPJ15610.1 KQ459249 KPJ02371.1 ODYU01011225 SOQ56835.1 KZ149958 PZC76437.1 AGBW02014444 OWR41661.1 GALX01003431 JAB65035.1 GEDC01019335 JAS17963.1 NEVH01009067 PNF34068.1 PNF34073.1 PNF34069.1 NWSH01003639 PCG66004.1 GEDC01003093 JAS34205.1 GBGD01001784 JAC87105.1 PNF34070.1 PNF34074.1 GEBQ01029891 GEBQ01004774 JAT10086.1 JAT35203.1 GFTR01005670 JAW10756.1 GECL01002701 JAP03423.1 GEBQ01016908 JAT23069.1 GEBQ01021655 JAT18322.1 PNF34075.1 GEBQ01004179 JAT35798.1 GEDC01007749 JAS29549.1 GALX01003432 JAB65034.1 PNF34072.1 GL732537 EFX83515.1 NNAY01000961 OXU25718.1 GDIQ01247799 GDIQ01242149 GDIP01098350 JAK03926.1 JAM05365.1 GDIQ01247797 GDIQ01242150 JAK03928.1 DS235088 EEB11623.1 GEZM01060854 JAV70850.1 KQ434796 KZC05786.1 GDIP01222594 GDIQ01247798 GDIQ01242148 GDIQ01024467 JAJ00808.1 JAK09577.1 KQ435711 KOX79573.1 KQ762882 OAD55341.1 GDIQ01254796 GDIQ01204110 GDIQ01192003 GDIP01098351 JAJ96928.1 JAM05364.1 GDIP01213744 JAJ09658.1 GBHO01017292 JAG26312.1 KQ971362 EFA08909.2 APGK01056920 APGK01056921 KB741277 KB632155 ENN71205.1 ERL89186.1 UFQS01000174 UFQT01000174 SSX00720.1 SSX21100.1 GBHO01017296 JAG26308.1 GEZM01060853 JAV70851.1 PCG66003.1 GDHC01008677 JAQ09952.1 GBHO01017297 JAG26307.1 GDIP01098349 GDIQ01070520 JAM05366.1 JAN24217.1 GDIQ01204111 JAK47614.1 GDIP01172093 JAJ51309.1 GL437468 EFN70415.1 JTDY01000361 KOB77541.1 GDIP01115108 GDIQ01021098 JAL88606.1 JAN73639.1 SSX00722.1 SSX21102.1 GDIQ01164932 JAK86793.1 ADTU01025150 ADTU01025151 ADTU01025152 ADTU01025153 GAKP01016184 JAC42768.1 GDIQ01098354 GDIP01138548 LRGB01002019 JAL53372.1 JAL65166.1 KZS09607.1 GAKP01016186 JAC42766.1 CVRI01000042 CRK95616.1 CRK95615.1 SSX00719.1 SSX21099.1 KK118212 KFM72467.1 GAKP01016185 GAKP01016183 JAC42769.1 KQ976619 KYM79092.1 GBXI01012754 JAD01538.1 GL888322 EGI62840.1 GAKP01016182 GAKP01016181 JAC42770.1 GDHF01021310 JAI31004.1 KQ982494 KYQ55728.1 KQ980653 KYN14805.1 GL764659 EFZ17327.1 KQ977808 KYM99543.1 IAAA01004547 LAA03376.1 IAAA01004548 LAA03379.1 JH430738 GFDF01001488 JAV12596.1 AJVK01007795 AJWK01030819 AJWK01030820 AJWK01030821
BAM19258.1 KQ460366 KPJ15610.1 KQ459249 KPJ02371.1 ODYU01011225 SOQ56835.1 KZ149958 PZC76437.1 AGBW02014444 OWR41661.1 GALX01003431 JAB65035.1 GEDC01019335 JAS17963.1 NEVH01009067 PNF34068.1 PNF34073.1 PNF34069.1 NWSH01003639 PCG66004.1 GEDC01003093 JAS34205.1 GBGD01001784 JAC87105.1 PNF34070.1 PNF34074.1 GEBQ01029891 GEBQ01004774 JAT10086.1 JAT35203.1 GFTR01005670 JAW10756.1 GECL01002701 JAP03423.1 GEBQ01016908 JAT23069.1 GEBQ01021655 JAT18322.1 PNF34075.1 GEBQ01004179 JAT35798.1 GEDC01007749 JAS29549.1 GALX01003432 JAB65034.1 PNF34072.1 GL732537 EFX83515.1 NNAY01000961 OXU25718.1 GDIQ01247799 GDIQ01242149 GDIP01098350 JAK03926.1 JAM05365.1 GDIQ01247797 GDIQ01242150 JAK03928.1 DS235088 EEB11623.1 GEZM01060854 JAV70850.1 KQ434796 KZC05786.1 GDIP01222594 GDIQ01247798 GDIQ01242148 GDIQ01024467 JAJ00808.1 JAK09577.1 KQ435711 KOX79573.1 KQ762882 OAD55341.1 GDIQ01254796 GDIQ01204110 GDIQ01192003 GDIP01098351 JAJ96928.1 JAM05364.1 GDIP01213744 JAJ09658.1 GBHO01017292 JAG26312.1 KQ971362 EFA08909.2 APGK01056920 APGK01056921 KB741277 KB632155 ENN71205.1 ERL89186.1 UFQS01000174 UFQT01000174 SSX00720.1 SSX21100.1 GBHO01017296 JAG26308.1 GEZM01060853 JAV70851.1 PCG66003.1 GDHC01008677 JAQ09952.1 GBHO01017297 JAG26307.1 GDIP01098349 GDIQ01070520 JAM05366.1 JAN24217.1 GDIQ01204111 JAK47614.1 GDIP01172093 JAJ51309.1 GL437468 EFN70415.1 JTDY01000361 KOB77541.1 GDIP01115108 GDIQ01021098 JAL88606.1 JAN73639.1 SSX00722.1 SSX21102.1 GDIQ01164932 JAK86793.1 ADTU01025150 ADTU01025151 ADTU01025152 ADTU01025153 GAKP01016184 JAC42768.1 GDIQ01098354 GDIP01138548 LRGB01002019 JAL53372.1 JAL65166.1 KZS09607.1 GAKP01016186 JAC42766.1 CVRI01000042 CRK95616.1 CRK95615.1 SSX00719.1 SSX21099.1 KK118212 KFM72467.1 GAKP01016185 GAKP01016183 JAC42769.1 KQ976619 KYM79092.1 GBXI01012754 JAD01538.1 GL888322 EGI62840.1 GAKP01016182 GAKP01016181 JAC42770.1 GDHF01021310 JAI31004.1 KQ982494 KYQ55728.1 KQ980653 KYN14805.1 GL764659 EFZ17327.1 KQ977808 KYM99543.1 IAAA01004547 LAA03376.1 IAAA01004548 LAA03379.1 JH430738 GFDF01001488 JAV12596.1 AJVK01007795 AJWK01030819 AJWK01030820 AJWK01030821
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000235965
UP000218220
+ More
UP000079169 UP000192223 UP000000305 UP000215335 UP000009046 UP000076502 UP000053105 UP000007266 UP000019118 UP000030742 UP000002358 UP000000311 UP000037510 UP000005203 UP000005205 UP000076858 UP000183832 UP000054359 UP000078540 UP000007755 UP000075809 UP000078492 UP000078542 UP000092462 UP000092461
UP000079169 UP000192223 UP000000305 UP000215335 UP000009046 UP000076502 UP000053105 UP000007266 UP000019118 UP000030742 UP000002358 UP000000311 UP000037510 UP000005203 UP000005205 UP000076858 UP000183832 UP000054359 UP000078540 UP000007755 UP000075809 UP000078492 UP000078542 UP000092462 UP000092461
Pfam
PF01513 NAD_kinase
Interpro
SUPFAM
SSF111331
SSF111331
Gene 3D
ProteinModelPortal
H9JD52
W8FNS9
W8FPH0
I4DMW8
A0A0N1PJB8
A0A194QFZ6
+ More
A0A2H1WUU3 A0A2W1BTA1 A0A212EJL7 V5G4S7 A0A1B6CXK4 A0A2J7QZQ3 A0A2J7QZR9 A0A2J7QZQ2 A0A2A4J3B5 A0A1B6E8I6 A0A069DT24 A0A2J7QZQ5 A0A2J7QZT2 A0A1B6MH08 A0A224XE60 A0A0V0G7T0 A0A1S3CYN3 A0A1B6LHG2 A0A1B6L3K8 A0A2J7QZS9 A0A1B6MIM4 A0A1B6DV81 A0A1S3CZZ4 A0A1S3CYP3 A0A1S3CYN8 A0A1S3CYM6 V5GLU2 A0A1S3CYM1 A0A1S3CYT3 A0A3Q0IQ79 A0A1S3CYQ9 A0A2J7QZQ7 A0A3Q0IQ14 A0A1S3CYL6 A0A1S3CYQ4 A0A1W4WVS0 A0A1S3CYS8 E9GAJ9 A0A232F5G1 A0A0P5G0Z2 A0A0P5HPC8 E0VE17 A0A1Y1LES9 A0A154P1H9 A0A0P5GI57 A0A0M9AAZ6 A0A310SKZ1 A0A0N8CU87 A0A0P4ZG78 A0A1W4X6L1 A0A0A9XZS4 D6WXV3 N6TPP3 A0A336LT55 A0A0A9Y5D0 K7ING6 A0A1Y1LD81 A0A2A4J3M7 A0A146LT56 A0A0A9XZR9 A0A0P6DZX2 A0A0P5JJL6 A0A0P5CEN9 E2A820 A0A0L7LQR6 A0A0P6HD38 A0A336KA94 A0A0P5LUN3 A0A088AM99 A0A158NST4 A0A034VML6 A0A0P5S5X8 A0A034VHK1 A0A1J1I5R3 A0A1J1I5K4 A0A336LWI5 A0A087U528 A0A034VJX2 A0A151I062 A0A158NST3 A0A0A1WSZ1 F4WSH5 A0A034VLW0 A0A0K8UWE6 A0A151X623 A0A195DPF4 E9IQ04 A0A151IF27 A0A2L2Y5B2 A0A2L2Y5E5 T1IL69 A0A1L8E1S5 A0A1B0GQR1 A0A1B0GKN9
A0A2H1WUU3 A0A2W1BTA1 A0A212EJL7 V5G4S7 A0A1B6CXK4 A0A2J7QZQ3 A0A2J7QZR9 A0A2J7QZQ2 A0A2A4J3B5 A0A1B6E8I6 A0A069DT24 A0A2J7QZQ5 A0A2J7QZT2 A0A1B6MH08 A0A224XE60 A0A0V0G7T0 A0A1S3CYN3 A0A1B6LHG2 A0A1B6L3K8 A0A2J7QZS9 A0A1B6MIM4 A0A1B6DV81 A0A1S3CZZ4 A0A1S3CYP3 A0A1S3CYN8 A0A1S3CYM6 V5GLU2 A0A1S3CYM1 A0A1S3CYT3 A0A3Q0IQ79 A0A1S3CYQ9 A0A2J7QZQ7 A0A3Q0IQ14 A0A1S3CYL6 A0A1S3CYQ4 A0A1W4WVS0 A0A1S3CYS8 E9GAJ9 A0A232F5G1 A0A0P5G0Z2 A0A0P5HPC8 E0VE17 A0A1Y1LES9 A0A154P1H9 A0A0P5GI57 A0A0M9AAZ6 A0A310SKZ1 A0A0N8CU87 A0A0P4ZG78 A0A1W4X6L1 A0A0A9XZS4 D6WXV3 N6TPP3 A0A336LT55 A0A0A9Y5D0 K7ING6 A0A1Y1LD81 A0A2A4J3M7 A0A146LT56 A0A0A9XZR9 A0A0P6DZX2 A0A0P5JJL6 A0A0P5CEN9 E2A820 A0A0L7LQR6 A0A0P6HD38 A0A336KA94 A0A0P5LUN3 A0A088AM99 A0A158NST4 A0A034VML6 A0A0P5S5X8 A0A034VHK1 A0A1J1I5R3 A0A1J1I5K4 A0A336LWI5 A0A087U528 A0A034VJX2 A0A151I062 A0A158NST3 A0A0A1WSZ1 F4WSH5 A0A034VLW0 A0A0K8UWE6 A0A151X623 A0A195DPF4 E9IQ04 A0A151IF27 A0A2L2Y5B2 A0A2L2Y5E5 T1IL69 A0A1L8E1S5 A0A1B0GQR1 A0A1B0GKN9
PDB
3PFN
E-value=5.13851e-110,
Score=1016
Ontologies
PATHWAY
GO
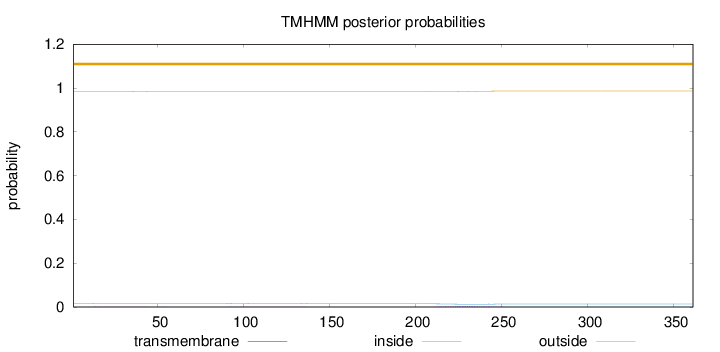
Topology
Length:
361
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13254
Exp number, first 60 AAs:
0.01671
Total prob of N-in:
0.01640
outside
1 - 361
Population Genetic Test Statistics
Pi
268.728462
Theta
204.337016
Tajima's D
0.909251
CLR
2.427746
CSRT
0.635168241587921
Interpretation
Uncertain
