Gene
KWMTBOMO01170 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007453
Annotation
Mn_superoxide_dismutase_[Bombyx_mori]
Full name
Superoxide dismutase
Location in the cell
Mitochondrial Reliability : 2.93
Sequence
CDS
ATGTCACAAAGGATTGGATCATTGATTCGAGTTGCCGGTGCTTCTCGCCAGAAGCATACTTTGCCAGAGCTTCCGTACGAGTACAATGCACTGGAGCCGGTCATTAGCCGTGAAATCATGAGTCTTCATCACAGCAAACATCACGCGACTTATATCAACAATCTAAACGTTGCTGAAGAGAAACTCGCACAGGCACAAGCTAAAGGTGATATCGACACCATTATCAACCTTGCACCAGCCTTGAAATTCAATGGTGGTGGTCACATCAACCACTCGATCTTTTGGCACAACCTGTCACCAAATGGTGGCAAGCCTTCTGATGTTCTCACCAAAGCCGTTGAGAAAGACTTTGGATCCTGGGATAACTTAAAGAATCAACTGTCGACAGCTTCTGTGGCAGTACAGGGCTCAGGCTGGGGTTGGCTTGGCTACAACAAACAAATGAAGAAATTGCAAATAGCTACATGCCAGAACCAGGATCCTCTGCAGGCCACCACTGGATTGGTCCCGCTCTTCGGAATCGATGTATGGGAGCACGCGTACTATCTTCAGTACAAGAACGTTCGTGCCGACTACGTGAAAGCTATTTTCGATGTAGCCAACTGGAATGATATATCTCAGAGATATGAAAAAGCGCTCAAGTGA
Protein
MSQRIGSLIRVAGASRQKHTLPELPYEYNALEPVISREIMSLHHSKHHATYINNLNVAEEKLAQAQAKGDIDTIINLAPALKFNGGGHINHSIFWHNLSPNGGKPSDVLTKAVEKDFGSWDNLKNQLSTASVAVQGSGWGWLGYNKQMKKLQIATCQNQDPLQATTGLVPLFGIDVWEHAYYLQYKNVRADYVKAIFDVANWNDISQRYEKALK
Summary
Description
Destroys radicals which are normally produced within the cells and which are toxic to biological systems.
Catalytic Activity
2 H(+) + 2 superoxide = H2O2 + O2
Similarity
Belongs to the iron/manganese superoxide dismutase family.
Feature
chain Superoxide dismutase
Uniprot
Q65Y02
H9JD56
A0A2Z4N450
A0A0H3WLT1
A0A2S0RQT9
A0A0N0PD16
+ More
A0A2A4ISP3 A0A0K0KJP6 A5Z1R7 A0A2W1BXF6 A0A212EU89 A0A2S0RQQ5 S4P9M2 D1M6Z4 A1E9A8 A0A067RBP2 A0A2P0XIZ9 A0A0H3WJR5 A0A3G9D4G0 A2SUH5 A7UL75 T2FE54 Q3Y596 K7IT33 A0A232FL74 G0Z047 M9T285 M4QL80 E6YCY4 D3J1L2 A0A2J7PDG3 E6YCX8 E6YCY7 A0A0P4WJN8 I6VM78 B9VUA3 I3VPK0 Q9NB66 E6YCX7 A0A0C9RVL9 A0A2A3EJB2 E6YCY2 E6YCY0 E6YCY5 E6YCY3 E6YCX9 A0A088A1K0 S4S5N4 Q7YXM5 E6YCY9 A0A0L7QNL3 E6YCY1 E6YCY8 A0A154PPR8 E1ZX89 E2B9W8 A0A0C9R9C6 H1ZSY8 R7V502 E6YCY6 A0A3M6TVI6 D6WWA9 A0A026WLL5 T1G7C2 A0A166AJ53 H2B644 E9IG44 F8R8R3 A0A087UP78 A0A2B4RVE0 A0A2I2MNA6 E1CPV0 A0A1B6KIM9 A0A2I2MNA9 A0A2I2MNB0 B4JWP5 A0A0K0EMH0 C3YI11 A0A195C2Y0 A0A226DD29 A0A0K0F4D5 J9XY23 A0A090L504 A0A2I2MNA3 A0A1B0DF57 A0A1Y1LRX0 A0A1B6FT12 A0A158NW84 A7RHP7 A0A0N5C416 A0A2P6LDK6 R4WD37 W8C1Z3 A0A1B6ICS9 W4Z933 A0A1I7S0V0 A0A183FRG8 A0A3P7YB45 A0A1L3THW9 A0A096X914
A0A2A4ISP3 A0A0K0KJP6 A5Z1R7 A0A2W1BXF6 A0A212EU89 A0A2S0RQQ5 S4P9M2 D1M6Z4 A1E9A8 A0A067RBP2 A0A2P0XIZ9 A0A0H3WJR5 A0A3G9D4G0 A2SUH5 A7UL75 T2FE54 Q3Y596 K7IT33 A0A232FL74 G0Z047 M9T285 M4QL80 E6YCY4 D3J1L2 A0A2J7PDG3 E6YCX8 E6YCY7 A0A0P4WJN8 I6VM78 B9VUA3 I3VPK0 Q9NB66 E6YCX7 A0A0C9RVL9 A0A2A3EJB2 E6YCY2 E6YCY0 E6YCY5 E6YCY3 E6YCX9 A0A088A1K0 S4S5N4 Q7YXM5 E6YCY9 A0A0L7QNL3 E6YCY1 E6YCY8 A0A154PPR8 E1ZX89 E2B9W8 A0A0C9R9C6 H1ZSY8 R7V502 E6YCY6 A0A3M6TVI6 D6WWA9 A0A026WLL5 T1G7C2 A0A166AJ53 H2B644 E9IG44 F8R8R3 A0A087UP78 A0A2B4RVE0 A0A2I2MNA6 E1CPV0 A0A1B6KIM9 A0A2I2MNA9 A0A2I2MNB0 B4JWP5 A0A0K0EMH0 C3YI11 A0A195C2Y0 A0A226DD29 A0A0K0F4D5 J9XY23 A0A090L504 A0A2I2MNA3 A0A1B0DF57 A0A1Y1LRX0 A0A1B6FT12 A0A158NW84 A7RHP7 A0A0N5C416 A0A2P6LDK6 R4WD37 W8C1Z3 A0A1B6ICS9 W4Z933 A0A1I7S0V0 A0A183FRG8 A0A3P7YB45 A0A1L3THW9 A0A096X914
EC Number
1.15.1.1
Pubmed
19121390
26354079
28756777
22118469
23622113
24845553
+ More
25700787 26760975 17097141 16621606 20075255 28648823 19835962 26394265 25366155 12769817 24269344 11932240 16139867 20798317 23254933 30382153 18362917 19820115 27387523 24508170 30249741 22062798 21282665 21565362 17994087 18563158 24120638 28004739 21347285 17615350 23691247 24495485 21909270 24905695
25700787 26760975 17097141 16621606 20075255 28648823 19835962 26394265 25366155 12769817 24269344 11932240 16139867 20798317 23254933 30382153 18362917 19820115 27387523 24508170 30249741 22062798 21282665 21565362 17994087 18563158 24120638 28004739 21347285 17615350 23691247 24495485 21909270 24905695
EMBL
AY886742
AB190802
AAW78358.1
BAD51413.1
BABH01036287
BABH01036288
+ More
BABH01036289 MG431819 AWX63642.1 KP407133 AKM12418.1 MF370559 RSAL01000160 AWA45967.1 RVE45454.1 KQ460366 KPJ15604.1 NWSH01007921 PCG62751.1 KM215271 AIJ00868.1 EF611125 ABR08302.1 KZ149958 PZC76443.1 AGBW02012439 OWR45060.1 MF991881 AWA45975.1 GAIX01003709 JAA88851.1 GU115810 ACY70995.1 EF114291 ABL63640.1 KK852562 KDR21306.1 KY661320 AVA17407.1 KP099968 AKM12646.1 FX985483 BAX29643.1 DQ205424 ABB05539.1 EU077525 ABU55004.1 KF156268 AGV76057.1 DQ157765 AAZ81617.1 AAZX01008089 NNAY01000061 OXU31412.1 HQ852225 AEK77428.1 KC461130 AGI99530.1 KC333178 AGH30393.1 FM213476 CAR82602.1 GQ478988 ADB90402.1 NEVH01026389 PNF14357.1 FM213470 CAR82596.1 FM213479 CAR82605.1 GDRN01000147 JAI68014.1 JX101466 AFN29184.1 FJ605170 JX133232 ACM61856.2 AFP89582.1 JQ763321 AFK82511.1 AF264029 AAF74770.1 FM213469 CAR82595.1 GBYB01013005 GBYB01013007 JAG82772.1 JAG82774.1 KZ288241 PBC31279.1 FM213474 CAR82600.1 FM213472 CAR82598.1 FM213477 CAR82603.1 FM213475 CAR82601.1 FM213471 CAR82597.1 JN637476 JN662941 AET74062.1 AET74063.1 AY329356 AAP93582.1 FM213481 CAR82607.1 KQ414851 KOC60215.1 FM213473 CAR82599.1 FM213480 CAR82606.1 KQ435012 KZC13862.1 GL435030 EFN74179.1 GL446605 EFN87555.1 GBYB01013004 JAG82771.1 FN652905 CBJ19215.1 AMQN01000856 KB296703 ELU11436.1 FM213478 CAR82604.1 RCHS01002830 RMX45412.1 LC154964 KQ971361 BAV32138.1 EFA09191.1 KK107154 QOIP01000001 EZA56942.1 RLU26398.1 AMQM01007545 KB097640 ESN92464.1 KU057379 AMZ80133.2 HQ115573 KF516615 ADV59552.1 AII16519.1 GL762910 EFZ20441.1 HM563680 ADM64421.1 KK120839 KFM79167.1 LSMT01000323 PFX20288.1 KU323377 AQQ11197.1 AB591738 BAJ21358.1 GEBQ01028667 JAT11310.1 KU323381 AQQ11201.1 KU323379 KU323380 KU323382 AQQ11199.1 AQQ11200.1 AQQ11202.1 CH916375 EDV98383.1 GG666514 EEN60148.1 KQ978317 KYM95219.1 LNIX01000022 OXA43475.1 JX402922 KU323368 KU323369 KU323370 KU323371 KU323372 KU323373 KU323375 KU323376 KU323378 AFS33106.1 AQQ11188.1 AQQ11189.1 AQQ11190.1 AQQ11191.1 AQQ11192.1 AQQ11193.1 AQQ11195.1 AQQ11196.1 AQQ11198.1 LN609528 CEF64881.1 KU323374 AQQ11194.1 AJVK01058555 GEZM01050447 JAV75588.1 GECZ01016439 JAS53330.1 ADTU01027848 DS469511 EDO49016.1 MWRG01000250 PRD36653.1 AK417363 BAN20578.1 GAMC01003352 JAC03204.1 GECU01022975 JAS84731.1 AAGJ04088089 UZAH01026761 VDO85026.1 KX695149 APH81360.1 KF471137 AHE13889.1
BABH01036289 MG431819 AWX63642.1 KP407133 AKM12418.1 MF370559 RSAL01000160 AWA45967.1 RVE45454.1 KQ460366 KPJ15604.1 NWSH01007921 PCG62751.1 KM215271 AIJ00868.1 EF611125 ABR08302.1 KZ149958 PZC76443.1 AGBW02012439 OWR45060.1 MF991881 AWA45975.1 GAIX01003709 JAA88851.1 GU115810 ACY70995.1 EF114291 ABL63640.1 KK852562 KDR21306.1 KY661320 AVA17407.1 KP099968 AKM12646.1 FX985483 BAX29643.1 DQ205424 ABB05539.1 EU077525 ABU55004.1 KF156268 AGV76057.1 DQ157765 AAZ81617.1 AAZX01008089 NNAY01000061 OXU31412.1 HQ852225 AEK77428.1 KC461130 AGI99530.1 KC333178 AGH30393.1 FM213476 CAR82602.1 GQ478988 ADB90402.1 NEVH01026389 PNF14357.1 FM213470 CAR82596.1 FM213479 CAR82605.1 GDRN01000147 JAI68014.1 JX101466 AFN29184.1 FJ605170 JX133232 ACM61856.2 AFP89582.1 JQ763321 AFK82511.1 AF264029 AAF74770.1 FM213469 CAR82595.1 GBYB01013005 GBYB01013007 JAG82772.1 JAG82774.1 KZ288241 PBC31279.1 FM213474 CAR82600.1 FM213472 CAR82598.1 FM213477 CAR82603.1 FM213475 CAR82601.1 FM213471 CAR82597.1 JN637476 JN662941 AET74062.1 AET74063.1 AY329356 AAP93582.1 FM213481 CAR82607.1 KQ414851 KOC60215.1 FM213473 CAR82599.1 FM213480 CAR82606.1 KQ435012 KZC13862.1 GL435030 EFN74179.1 GL446605 EFN87555.1 GBYB01013004 JAG82771.1 FN652905 CBJ19215.1 AMQN01000856 KB296703 ELU11436.1 FM213478 CAR82604.1 RCHS01002830 RMX45412.1 LC154964 KQ971361 BAV32138.1 EFA09191.1 KK107154 QOIP01000001 EZA56942.1 RLU26398.1 AMQM01007545 KB097640 ESN92464.1 KU057379 AMZ80133.2 HQ115573 KF516615 ADV59552.1 AII16519.1 GL762910 EFZ20441.1 HM563680 ADM64421.1 KK120839 KFM79167.1 LSMT01000323 PFX20288.1 KU323377 AQQ11197.1 AB591738 BAJ21358.1 GEBQ01028667 JAT11310.1 KU323381 AQQ11201.1 KU323379 KU323380 KU323382 AQQ11199.1 AQQ11200.1 AQQ11202.1 CH916375 EDV98383.1 GG666514 EEN60148.1 KQ978317 KYM95219.1 LNIX01000022 OXA43475.1 JX402922 KU323368 KU323369 KU323370 KU323371 KU323372 KU323373 KU323375 KU323376 KU323378 AFS33106.1 AQQ11188.1 AQQ11189.1 AQQ11190.1 AQQ11191.1 AQQ11192.1 AQQ11193.1 AQQ11195.1 AQQ11196.1 AQQ11198.1 LN609528 CEF64881.1 KU323374 AQQ11194.1 AJVK01058555 GEZM01050447 JAV75588.1 GECZ01016439 JAS53330.1 ADTU01027848 DS469511 EDO49016.1 MWRG01000250 PRD36653.1 AK417363 BAN20578.1 GAMC01003352 JAC03204.1 GECU01022975 JAS84731.1 AAGJ04088089 UZAH01026761 VDO85026.1 KX695149 APH81360.1 KF471137 AHE13889.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000218220
UP000007151
UP000027135
+ More
UP000002358 UP000215335 UP000235965 UP000242457 UP000005203 UP000053825 UP000076502 UP000000311 UP000008237 UP000014760 UP000275408 UP000007266 UP000053097 UP000279307 UP000015101 UP000054359 UP000225706 UP000001070 UP000035681 UP000001554 UP000078542 UP000198287 UP000035680 UP000035682 UP000092462 UP000005205 UP000001593 UP000046392 UP000007110 UP000095284 UP000050761
UP000002358 UP000215335 UP000235965 UP000242457 UP000005203 UP000053825 UP000076502 UP000000311 UP000008237 UP000014760 UP000275408 UP000007266 UP000053097 UP000279307 UP000015101 UP000054359 UP000225706 UP000001070 UP000035681 UP000001554 UP000078542 UP000198287 UP000035680 UP000035682 UP000092462 UP000005205 UP000001593 UP000046392 UP000007110 UP000095284 UP000050761
Interpro
Gene 3D
ProteinModelPortal
Q65Y02
H9JD56
A0A2Z4N450
A0A0H3WLT1
A0A2S0RQT9
A0A0N0PD16
+ More
A0A2A4ISP3 A0A0K0KJP6 A5Z1R7 A0A2W1BXF6 A0A212EU89 A0A2S0RQQ5 S4P9M2 D1M6Z4 A1E9A8 A0A067RBP2 A0A2P0XIZ9 A0A0H3WJR5 A0A3G9D4G0 A2SUH5 A7UL75 T2FE54 Q3Y596 K7IT33 A0A232FL74 G0Z047 M9T285 M4QL80 E6YCY4 D3J1L2 A0A2J7PDG3 E6YCX8 E6YCY7 A0A0P4WJN8 I6VM78 B9VUA3 I3VPK0 Q9NB66 E6YCX7 A0A0C9RVL9 A0A2A3EJB2 E6YCY2 E6YCY0 E6YCY5 E6YCY3 E6YCX9 A0A088A1K0 S4S5N4 Q7YXM5 E6YCY9 A0A0L7QNL3 E6YCY1 E6YCY8 A0A154PPR8 E1ZX89 E2B9W8 A0A0C9R9C6 H1ZSY8 R7V502 E6YCY6 A0A3M6TVI6 D6WWA9 A0A026WLL5 T1G7C2 A0A166AJ53 H2B644 E9IG44 F8R8R3 A0A087UP78 A0A2B4RVE0 A0A2I2MNA6 E1CPV0 A0A1B6KIM9 A0A2I2MNA9 A0A2I2MNB0 B4JWP5 A0A0K0EMH0 C3YI11 A0A195C2Y0 A0A226DD29 A0A0K0F4D5 J9XY23 A0A090L504 A0A2I2MNA3 A0A1B0DF57 A0A1Y1LRX0 A0A1B6FT12 A0A158NW84 A7RHP7 A0A0N5C416 A0A2P6LDK6 R4WD37 W8C1Z3 A0A1B6ICS9 W4Z933 A0A1I7S0V0 A0A183FRG8 A0A3P7YB45 A0A1L3THW9 A0A096X914
A0A2A4ISP3 A0A0K0KJP6 A5Z1R7 A0A2W1BXF6 A0A212EU89 A0A2S0RQQ5 S4P9M2 D1M6Z4 A1E9A8 A0A067RBP2 A0A2P0XIZ9 A0A0H3WJR5 A0A3G9D4G0 A2SUH5 A7UL75 T2FE54 Q3Y596 K7IT33 A0A232FL74 G0Z047 M9T285 M4QL80 E6YCY4 D3J1L2 A0A2J7PDG3 E6YCX8 E6YCY7 A0A0P4WJN8 I6VM78 B9VUA3 I3VPK0 Q9NB66 E6YCX7 A0A0C9RVL9 A0A2A3EJB2 E6YCY2 E6YCY0 E6YCY5 E6YCY3 E6YCX9 A0A088A1K0 S4S5N4 Q7YXM5 E6YCY9 A0A0L7QNL3 E6YCY1 E6YCY8 A0A154PPR8 E1ZX89 E2B9W8 A0A0C9R9C6 H1ZSY8 R7V502 E6YCY6 A0A3M6TVI6 D6WWA9 A0A026WLL5 T1G7C2 A0A166AJ53 H2B644 E9IG44 F8R8R3 A0A087UP78 A0A2B4RVE0 A0A2I2MNA6 E1CPV0 A0A1B6KIM9 A0A2I2MNA9 A0A2I2MNB0 B4JWP5 A0A0K0EMH0 C3YI11 A0A195C2Y0 A0A226DD29 A0A0K0F4D5 J9XY23 A0A090L504 A0A2I2MNA3 A0A1B0DF57 A0A1Y1LRX0 A0A1B6FT12 A0A158NW84 A7RHP7 A0A0N5C416 A0A2P6LDK6 R4WD37 W8C1Z3 A0A1B6ICS9 W4Z933 A0A1I7S0V0 A0A183FRG8 A0A3P7YB45 A0A1L3THW9 A0A096X914
PDB
3DC6
E-value=1.84586e-76,
Score=724
Ontologies
PATHWAY
GO
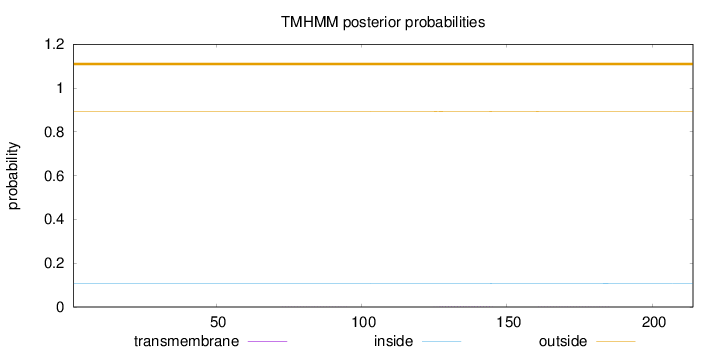
Topology
Length:
214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06295
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10627
outside
1 - 214
Population Genetic Test Statistics
Pi
194.417121
Theta
223.126563
Tajima's D
-0.405172
CLR
0.614531
CSRT
0.267936603169842
Interpretation
Uncertain
