Gene
KWMTBOMO01167
Pre Gene Modal
BGIBMGA007455
Annotation
PREDICTED:_kinesin-like_protein_unc-104_isoform_X3_[Bombyx_mori]
Full name
Kinesin-like protein unc-104
Alternative Name
Protein immaculate connections
Location in the cell
Cytoplasmic Reliability : 2.429
Sequence
CDS
ATGGTGTACAAAGACATCGGTGAAGAGATGTTACAACACGCTTTCGACGGTTACAATATATGCATATTCGCTTACGGTCAGACCGGGGCTGGTAAATCTTATACGATGATGGGCCGCGGTGAGGATGGTCAGGAGGGCATCATTCCTCAGATCTGCAAGGATCTGTTCAGGCGCATCAGACAGACTACGTCCGATGATCTCAAATATTCGGTTGAAGTGTCGTACATGGAAATTTATTGCGAACGGGTTCGTGACTTACTGAACCCTAAGAATAAGGGAAATCTTCGGGTCCGTGAGCACCCCGCCCTCGGACCGTACGTGGAGGATCTCAGCAAACTGGCGGTCACCTCCTACCAAGACATTTACGATCTCATCGACGAGGGTAACAAAGCTAGGACCGTGGCGGCTACGAACATGAACGAGACGTCGTCTCGCTCTCACGCCGTCTTCACCATATTCTTCACGCAGCAACGTCATGACGTCACTACGAATTTGATGTCCGAAAAGGTCTCGAAAATATCGCTTGTCGATCTGGCCGGCTCGGAGCGTGCCGATTCTACCGGAGCCAAAGGAACTCGTCTCAAGGAAGGAGCTAACATCAACAAATCTCTGACCACACTCGGAAAAGTGATTTCCGCTCTAGCCGAAATTGCCTCTAAGAGCAAGAAGTCCAAGAAGGCGGACTTCATACCGTACCGTGACTCGGTGCTGACGTGGCTGCTTCGCGAGAACCTCGGCGGTAACTCGAAGACCGCCATGATAGCCGCCATCTCGCCGGCCGACATCAACTTCGACGAGACGCTCAGTACACTTAGATACGCGGATCGAGCCAAACAGATCGTATGCAAGGCGATCGTGAACGAGGACGCGAACGCTAAACTCATTCGCGAACTCAAAGAGGAGATCCTGAAGCTTCGCGAACTGCTCAAGGCTGAAGGGATCGACGTGGAAGAAGGCGAACAAAATTCGTCGCCGCAACGCAAAAAGAGCGAAGCAGAGGTTCTGTCGCCGAAGCTGTCCCGAGCCGCCACCACCATCGCCGAGGAGGCAGTGGATCAGCTCCAAGCCTCCGAGAAGCTTATCGCTGAACTCAACGAAACTTGGGAGGAAAAACTCAAGCGGACAGAACAGATCCGCGTCCAGCGCGAAGCGGTCTTCGCGGAAATGGGTCTTGCTGTCAAGGAGGGCATCACCGTCGGCATATACTCGCCGAAGAAAACGCCACACCTCGTCAACTTGAACGAAGACCCGAACCTGTCCGAATGCCTGCTTTACTATATAAAAGACGGTGTGACTCGCGTGGGCACATCTGAAGCGAAAGTCCCACAAGACATCCAGCTGTCCGGCTCGCACATACTGAGCGAGCACTGCATCTTCGAGAACGCGGACGGCGTGATCTGCCTCATTCCGCACCGCGGGGCACTGGTCTACGTCAACGGACGGGAGCTGGAAGAGCCCGTGATACTGAAGAGCGGTTCCCGCGTGATCCTGGGCAAGAACCACGTGTTCCGGTTCACCCACCCCGGGCAGCCGCGCGAGCTGCCGCCCGTCAACAAGGCGCTCACAGACTCCACCGAGTCGGCAGCTGAAGTCGCTACCGAGAGCAGCAGCGATACAGAAAATCAGAACGTCGACTGGGACTTCGCTCAGTGCGAGCTGCTAGAGAAACAGGGCATCGACTTGAAGGCGGAGATGCAGAAGCGTCTGATGGCCCTCGAGGAACAGTTCCGTAGGGAGAAGGAGCACGCCGACCAGCAGTTTGAGGAGCAGCGTAAGAACTACGAGGCTCGCATCGATGCTCTCCAGCGTCAGGTGGAGGAGCAGAGCGTAACGATGTCCATGTACAGCTCCTACACGCCCGAAGACTTCCATAACGATGAAGATATATTCGTGAACCCGTTGTTCGAGACGGAGTGCTGGTCGGCTCGCGAGGTGGGGCTGGCGGCCTGGGCCTTCCGCAAGTGGAAGTACCATCAGTTCACGTCGCTCCGAGACGACCTCTGGGGAAATGCGATCTTCCTCAAGGAAGCGAACGCGATCTCTGTGGAGCTGCGCAAGAAAGTTCAGTTTCAGTTCACTCTGCTAACGGACACGCCGTACTCCCCCCTCCCCGCGGAGCTGGCCCCCCGCGACGACGCCGACGACGAGTACCGGCCCAGCGCGCCCACGGTGGTCGCCGTCGAGGTCACCGACACCAAGAACGGCGCCACGCACTACTGGACCCTAGAGAAGCTACGCCATCGTCTGGAGCTAATGCGTCAGATATACAACATGAACGAGAGCTCGTGCGTAGAGGACGAGGAGACCACGCCCCTAGGTGCTCCGCCCTCGATCGGGGCGCCGCCCCCGCCCGCCGACATCCTACAATGTATATCGGCCTGCGCGCAACAAACCCGCCTCTCCCTAGCCAACTTGCTACCATCGAGACAACGGCTCGAGTTAATGCGCGAGATGTATCACAACGAGGCCGAGCTGTCGCCTACTTCGCCCGACCACAACATCGAGTCGGTAACCGGCGGCGATCCTTTCTACGATAGGTTCCCGTGGTTCCGTCTTGTTGGACGGAGCTTTGTATACTTGTCGAACCTCCTGTACCCGGTTCCTCTCATTCACAAGGTCGCTATAGTGAACGAGAAAGGAGACGTGAAGGGCTACCTCCGGGTCGCCGTGCAGGCCGTTATCGACGCCGAGAAGAACAACGCGGAATTCGCAGCTGGCGTGAAGCAGTCAGCCAAGATATGCTTCGACGACGATGTAGTGCCGCGCGCCCGGGTCCGCGCCGCCAACAGCGTCGACAAGAACAACGCCATCAACCTGGACGAACGCGCCGTCGAGGTCCAGGACTCGAACATCAAAATTGAGGAATTGGCTGGAGGAGAGTGCGATGCGGATAGCGGCCGCGGGGACAGCTCTCTCGCTTCTGAACTGAAAGAGGAAGACTTGCCTGAACACTTGACCCTCGGCAAGGAGTTCACCTTCCGCGTCACCGTGCTCCAAGCTCACAGCGTCGCCACCGACTATGCTGATGTCTTCTGTCAGTTCAACTTCCTTCACCGCAACGAGGACGCCTTCTCCACCGAACCCGTCAAGAACGCAGGAAAAAATACACCACTCGGATTCTACCACGTGCAGAATATAACTGTACCGATTACAAAATCATTTGTGGAATACATTAAAACTCAACCGATCGTGTTTGAAGTCTTCGGTCATTATCAGCAGCATCCGTTACATAAGGATGCGAAACAAGATGGCCCTGTGGGAGGCCGCACGCCCCCGCGCCGCATGCTGCCGCCCTCCATCCCCATCTCGGCCCCCGTGCGGAGCCCCAAGTGGGGGGCCGCCAGTGTGGCGCCGCCCTGCTGCTCCTCGCACCTGCACTCCAAGCATGACCTGCTCGTCTGGTTCGAGATCTGCGAGCTCGCACCCAATGGAGAATACGTACCTGCCGTGGTGGAGCACAGTGACGAGCTCCCCTGCCGCGGGCTCTTCCTTCTGCACCAGGGAATCCAGCGCCGCATCCGCATCACCATCCTGCACGAGCCCTCCGCTGACCTGCAGTGGACCGATGTCAGGGAGCTCGTTGTCGGTCGCATCCGCAACACGCCCGAAGCCAATGAGGATACCACTGACGGGGACGAGGATGGCGCCCTCTCACTCGGCCTGTTCCCCGGGGAGAGACCCGCGCTCGACGATCGGGCCGTCTTCCGGTTCGAAGCGGCCTGGGACAGCAGCCTGCACGGCTCGCCGCTACTGAACCGAGTGAGCGCTAACGGGGAAGTGGTCTACATCACGCTCAGCGCTTACCTCGAGATGGAGAACTGCGGGCGGCCGGCGATCGTGACGAAGGACCTGTCGGTGGTGGTGGTAGGTCGCGAGGCGCGCACGGGCCGTTCGCTGCGCCGCGCACTGTTTGGGTCCCGCGCCGCCCGCGCCGACCACATCACCGGCGTCTACGAGCTCAGCCTGCACCGCGCGCTCGAACCCGGAGTCCAGCGCCGGCAGCGTCGCGTGCTGGACACGAGCGGCACGTACGTGCGCGGGGAAGAGAACCTACACGGGTGGAGGCCGCGCGGGGACTCGCTCATATTCGACCACCAGGTAATTACGCAAATTATCATTTTTCATGTTTGA
Protein
MVYKDIGEEMLQHAFDGYNICIFAYGQTGAGKSYTMMGRGEDGQEGIIPQICKDLFRRIRQTTSDDLKYSVEVSYMEIYCERVRDLLNPKNKGNLRVREHPALGPYVEDLSKLAVTSYQDIYDLIDEGNKARTVAATNMNETSSRSHAVFTIFFTQQRHDVTTNLMSEKVSKISLVDLAGSERADSTGAKGTRLKEGANINKSLTTLGKVISALAEIASKSKKSKKADFIPYRDSVLTWLLRENLGGNSKTAMIAAISPADINFDETLSTLRYADRAKQIVCKAIVNEDANAKLIRELKEEILKLRELLKAEGIDVEEGEQNSSPQRKKSEAEVLSPKLSRAATTIAEEAVDQLQASEKLIAELNETWEEKLKRTEQIRVQREAVFAEMGLAVKEGITVGIYSPKKTPHLVNLNEDPNLSECLLYYIKDGVTRVGTSEAKVPQDIQLSGSHILSEHCIFENADGVICLIPHRGALVYVNGRELEEPVILKSGSRVILGKNHVFRFTHPGQPRELPPVNKALTDSTESAAEVATESSSDTENQNVDWDFAQCELLEKQGIDLKAEMQKRLMALEEQFRREKEHADQQFEEQRKNYEARIDALQRQVEEQSVTMSMYSSYTPEDFHNDEDIFVNPLFETECWSAREVGLAAWAFRKWKYHQFTSLRDDLWGNAIFLKEANAISVELRKKVQFQFTLLTDTPYSPLPAELAPRDDADDEYRPSAPTVVAVEVTDTKNGATHYWTLEKLRHRLELMRQIYNMNESSCVEDEETTPLGAPPSIGAPPPPADILQCISACAQQTRLSLANLLPSRQRLELMREMYHNEAELSPTSPDHNIESVTGGDPFYDRFPWFRLVGRSFVYLSNLLYPVPLIHKVAIVNEKGDVKGYLRVAVQAVIDAEKNNAEFAAGVKQSAKICFDDDVVPRARVRAANSVDKNNAINLDERAVEVQDSNIKIEELAGGECDADSGRGDSSLASELKEEDLPEHLTLGKEFTFRVTVLQAHSVATDYADVFCQFNFLHRNEDAFSTEPVKNAGKNTPLGFYHVQNITVPITKSFVEYIKTQPIVFEVFGHYQQHPLHKDAKQDGPVGGRTPPRRMLPPSIPISAPVRSPKWGAASVAPPCCSSHLHSKHDLLVWFEICELAPNGEYVPAVVEHSDELPCRGLFLLHQGIQRRIRITILHEPSADLQWTDVRELVVGRIRNTPEANEDTTDGDEDGALSLGLFPGERPALDDRAVFRFEAAWDSSLHGSPLLNRVSANGEVVYITLSAYLEMENCGRPAIVTKDLSVVVVGREARTGRSLRRALFGSRAARADHITGVYELSLHRALEPGVQRRQRRVLDTSGTYVRGEENLHGWRPRGDSLIFDHQVITQIIIFHV
Summary
Description
Required for presynaptic maturation, has a role in axonal transport of dense-core vesicles carrying synaptic vesicle precursors, components required for the morphological transformation of axonal growth cones to mature boutons.
Subunit
Monomer (By similarity). Interacts with Arl8 (PubMed:30174114).
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. Unc-104 subfamily.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. Unc-104 subfamily.
Keywords
Alternative splicing
ATP-binding
Cell projection
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Microtubule
Motor protein
Nucleotide-binding
Phosphoprotein
Protein transport
Reference proteome
Transport
Feature
chain Kinesin-like protein unc-104
splice variant In isoform E.
splice variant In isoform E.
Uniprot
A0A2H1WUC8
A0A194QBJ1
A0A2J7Q5K3
A0A2J7Q5K0
A0A0C9RLS1
A0A088A1K2
+ More
E1ZX90 A0A2J7Q5K2 F4WBT9 A0A139WDX9 A0A2A3EHV1 A0A195C3P2 A0A151WN78 A0A195FU85 A0A3L8E154 A0A158NW85 A0A195BD68 D6WVN9 A0A0L7QNY7 A0A195DI52 E2B9W7 A0A336LU78 A0A336LUI3 A0A0M8ZQ99 B4MIT1 A0A0R1DSR7 B3NP74 A0A0R1DUM0 A0A0R1DSJ0 A0A084WAX1 A1ZAJ2-2 A0A0R1DY12 A0A0Q5W0R5 U4U2Y2 A0A0Q5VLD5 A0A0R1DSK4 A0A0R1DY25 A0A0Q5VL17 A0A0Q5VZM5 A0A0Q5VLV6 B3MDI4 B4QIF3 A0A0J9U3T7 A0A0J9RFJ1 N6W6Y3 A0A0M4EVV7 A0A182JSX9 A0A0J9RE31 A0A0J9RFJ7 N6SZ40 A0A0J9U3U8 A0A0R3NQ16 A0A0R3NV63 A0A1A9UHE4 A0A0R3NQ74 A0A0R3NWD4 A0A0R3NQT2 A0A0R3NVL4 A0A1A9Z4G3 A0A1A9WWP0 X1WXH4 A0A1B0BZT0 B0WDG3 A0A1B0CG12 T1IJB9 B4H744 A0A2C9M9P4 A0A2C9M9S1 A0A2C9M9R5 A0A2C9M9R1 A0A2C9M9R2 A0A2C9MA25 A0A2C9M9R0 A0A2C9MA21 A0A2C9M9S0 K1RN38 A0A087URW9 A0A0V1MPD1 A0A0V1MPB9 A0A0V1H0V9 A0A0V1H0T9 A0A0V1EG07 A0A0V1IIL9 A0A0V1ABV8 A0A0V1CHM6 A0A0V0RHW1 A0A0V0RHP6 A0A0V0V2J5 A0A0V1BA73 A0A0V0RHH0 A0A0V0TF06 A0A0V1BAF1 A0A0V0TG64 A0A0V1BAH8 A0A0V1PG02 A0A0V1PFS2 A0A0V1H109 A0A0V1BA88
E1ZX90 A0A2J7Q5K2 F4WBT9 A0A139WDX9 A0A2A3EHV1 A0A195C3P2 A0A151WN78 A0A195FU85 A0A3L8E154 A0A158NW85 A0A195BD68 D6WVN9 A0A0L7QNY7 A0A195DI52 E2B9W7 A0A336LU78 A0A336LUI3 A0A0M8ZQ99 B4MIT1 A0A0R1DSR7 B3NP74 A0A0R1DUM0 A0A0R1DSJ0 A0A084WAX1 A1ZAJ2-2 A0A0R1DY12 A0A0Q5W0R5 U4U2Y2 A0A0Q5VLD5 A0A0R1DSK4 A0A0R1DY25 A0A0Q5VL17 A0A0Q5VZM5 A0A0Q5VLV6 B3MDI4 B4QIF3 A0A0J9U3T7 A0A0J9RFJ1 N6W6Y3 A0A0M4EVV7 A0A182JSX9 A0A0J9RE31 A0A0J9RFJ7 N6SZ40 A0A0J9U3U8 A0A0R3NQ16 A0A0R3NV63 A0A1A9UHE4 A0A0R3NQ74 A0A0R3NWD4 A0A0R3NQT2 A0A0R3NVL4 A0A1A9Z4G3 A0A1A9WWP0 X1WXH4 A0A1B0BZT0 B0WDG3 A0A1B0CG12 T1IJB9 B4H744 A0A2C9M9P4 A0A2C9M9S1 A0A2C9M9R5 A0A2C9M9R1 A0A2C9M9R2 A0A2C9MA25 A0A2C9M9R0 A0A2C9MA21 A0A2C9M9S0 K1RN38 A0A087URW9 A0A0V1MPD1 A0A0V1MPB9 A0A0V1H0V9 A0A0V1H0T9 A0A0V1EG07 A0A0V1IIL9 A0A0V1ABV8 A0A0V1CHM6 A0A0V0RHW1 A0A0V0RHP6 A0A0V0V2J5 A0A0V1BA73 A0A0V0RHH0 A0A0V0TF06 A0A0V1BAF1 A0A0V0TG64 A0A0V1BAH8 A0A0V1PG02 A0A0V1PFS2 A0A0V1H109 A0A0V1BA88
Pubmed
EMBL
ODYU01011089
SOQ56607.1
KQ459249
KPJ02360.1
NEVH01017557
PNF23861.1
+ More
PNF23862.1 GBYB01007926 JAG77693.1 GL435030 EFN74180.1 PNF23863.1 GL888066 EGI68367.1 KQ971357 KYB26178.1 KZ288241 PBC31278.1 KQ978317 KYM95220.1 KQ982911 KYQ49316.1 KQ981280 KYN43449.1 QOIP01000001 RLU26397.1 ADTU01027849 ADTU01027850 KQ976511 KYM82516.1 EFA08595.1 KQ414851 KOC60216.1 KQ980824 KYN12573.1 GL446605 EFN87554.1 UFQT01000198 SSX21596.1 SSX21595.1 KQ435971 KOX67776.1 CH963719 EDW72020.2 CM000158 KRK00173.1 CH954179 EDV55713.1 KRK00171.1 KRK00174.1 KRK00176.1 ATLV01022273 KE525331 KFB47365.1 AF247761 AE013599 AAF74192.1 KRK00162.1 KQS62424.1 KB631948 ERL87402.1 KQS62416.1 KRK00168.1 KRK00172.1 KRK00177.1 KQS62413.1 KQS62418.1 KQS62420.1 KQS62423.1 CH902619 EDV37447.2 KPU76827.1 CM000362 CM002911 EDX07400.1 KMY94336.1 KMY94340.1 KMY94339.1 CM000071 ENO01988.1 CP012524 ALC42087.1 KMY94338.1 KMY94342.1 KMY94343.1 KMY94344.1 APGK01058543 KB741291 ENN70508.1 KMY94350.1 KRT03230.1 KRT03231.1 KRT03223.1 KRT03232.1 KRT03228.1 KRT03224.1 KRT03229.1 ABLF02019510 JXJN01023289 DS231897 EDS44534.1 AJWK01010636 AJWK01010637 AJWK01010638 AJWK01010639 JH430253 CH479216 EDW33577.1 JH818911 EKC43000.1 KK121271 KFM80108.1 JYDO01000060 KRZ73655.1 KRZ73652.1 JYDP01000171 KRZ04176.1 KRZ04178.1 JYDR01000043 KRY72570.1 JYDS01000186 JYDV01000073 KRZ22037.1 KRZ36424.1 JYDQ01000010 KRY22309.1 JYDI01000197 KRY48757.1 JYDL01000176 KRX13967.1 KRX13970.1 JYDN01000096 KRX57737.1 JYDH01000075 KRY33908.1 KRX13966.1 JYDJ01000305 KRX37537.1 KRY33906.1 KRX37539.1 KRY33905.1 JYDM01000014 KRZ95084.1 KRZ95085.1 KRZ04177.1 KRY33904.1
PNF23862.1 GBYB01007926 JAG77693.1 GL435030 EFN74180.1 PNF23863.1 GL888066 EGI68367.1 KQ971357 KYB26178.1 KZ288241 PBC31278.1 KQ978317 KYM95220.1 KQ982911 KYQ49316.1 KQ981280 KYN43449.1 QOIP01000001 RLU26397.1 ADTU01027849 ADTU01027850 KQ976511 KYM82516.1 EFA08595.1 KQ414851 KOC60216.1 KQ980824 KYN12573.1 GL446605 EFN87554.1 UFQT01000198 SSX21596.1 SSX21595.1 KQ435971 KOX67776.1 CH963719 EDW72020.2 CM000158 KRK00173.1 CH954179 EDV55713.1 KRK00171.1 KRK00174.1 KRK00176.1 ATLV01022273 KE525331 KFB47365.1 AF247761 AE013599 AAF74192.1 KRK00162.1 KQS62424.1 KB631948 ERL87402.1 KQS62416.1 KRK00168.1 KRK00172.1 KRK00177.1 KQS62413.1 KQS62418.1 KQS62420.1 KQS62423.1 CH902619 EDV37447.2 KPU76827.1 CM000362 CM002911 EDX07400.1 KMY94336.1 KMY94340.1 KMY94339.1 CM000071 ENO01988.1 CP012524 ALC42087.1 KMY94338.1 KMY94342.1 KMY94343.1 KMY94344.1 APGK01058543 KB741291 ENN70508.1 KMY94350.1 KRT03230.1 KRT03231.1 KRT03223.1 KRT03232.1 KRT03228.1 KRT03224.1 KRT03229.1 ABLF02019510 JXJN01023289 DS231897 EDS44534.1 AJWK01010636 AJWK01010637 AJWK01010638 AJWK01010639 JH430253 CH479216 EDW33577.1 JH818911 EKC43000.1 KK121271 KFM80108.1 JYDO01000060 KRZ73655.1 KRZ73652.1 JYDP01000171 KRZ04176.1 KRZ04178.1 JYDR01000043 KRY72570.1 JYDS01000186 JYDV01000073 KRZ22037.1 KRZ36424.1 JYDQ01000010 KRY22309.1 JYDI01000197 KRY48757.1 JYDL01000176 KRX13967.1 KRX13970.1 JYDN01000096 KRX57737.1 JYDH01000075 KRY33908.1 KRX13966.1 JYDJ01000305 KRX37537.1 KRY33906.1 KRX37539.1 KRY33905.1 JYDM01000014 KRZ95084.1 KRZ95085.1 KRZ04177.1 KRY33904.1
Proteomes
UP000053268
UP000235965
UP000005203
UP000000311
UP000007755
UP000007266
+ More
UP000242457 UP000078542 UP000075809 UP000078541 UP000279307 UP000005205 UP000078540 UP000053825 UP000078492 UP000008237 UP000053105 UP000007798 UP000002282 UP000008711 UP000030765 UP000000803 UP000030742 UP000007801 UP000000304 UP000001819 UP000092553 UP000075881 UP000019118 UP000078200 UP000092445 UP000091820 UP000007819 UP000092460 UP000002320 UP000092461 UP000008744 UP000076420 UP000005408 UP000054359 UP000054843 UP000055024 UP000054632 UP000054805 UP000054826 UP000054783 UP000054653 UP000054630 UP000054681 UP000054776 UP000055048 UP000054924
UP000242457 UP000078542 UP000075809 UP000078541 UP000279307 UP000005205 UP000078540 UP000053825 UP000078492 UP000008237 UP000053105 UP000007798 UP000002282 UP000008711 UP000030765 UP000000803 UP000030742 UP000007801 UP000000304 UP000001819 UP000092553 UP000075881 UP000019118 UP000078200 UP000092445 UP000091820 UP000007819 UP000092460 UP000002320 UP000092461 UP000008744 UP000076420 UP000005408 UP000054359 UP000054843 UP000055024 UP000054632 UP000054805 UP000054826 UP000054783 UP000054653 UP000054630 UP000054681 UP000054776 UP000055048 UP000054924
PRIDE
Pfam
Interpro
IPR008984
SMAD_FHA_dom_sf
+ More
IPR000253 FHA_dom
IPR022164 Kinesin-like
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR001849 PH_domain
IPR011993 PH-like_dom_sf
IPR036961 Kinesin_motor_dom_sf
IPR032405 Kinesin_assoc
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR022140 Kinesin-like_KIF1-typ
IPR038765 Papain-like_cys_pep_sf
IPR003323 OTU_dom
IPR000253 FHA_dom
IPR022164 Kinesin-like
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR001849 PH_domain
IPR011993 PH-like_dom_sf
IPR036961 Kinesin_motor_dom_sf
IPR032405 Kinesin_assoc
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR022140 Kinesin-like_KIF1-typ
IPR038765 Papain-like_cys_pep_sf
IPR003323 OTU_dom
Gene 3D
CDD
ProteinModelPortal
A0A2H1WUC8
A0A194QBJ1
A0A2J7Q5K3
A0A2J7Q5K0
A0A0C9RLS1
A0A088A1K2
+ More
E1ZX90 A0A2J7Q5K2 F4WBT9 A0A139WDX9 A0A2A3EHV1 A0A195C3P2 A0A151WN78 A0A195FU85 A0A3L8E154 A0A158NW85 A0A195BD68 D6WVN9 A0A0L7QNY7 A0A195DI52 E2B9W7 A0A336LU78 A0A336LUI3 A0A0M8ZQ99 B4MIT1 A0A0R1DSR7 B3NP74 A0A0R1DUM0 A0A0R1DSJ0 A0A084WAX1 A1ZAJ2-2 A0A0R1DY12 A0A0Q5W0R5 U4U2Y2 A0A0Q5VLD5 A0A0R1DSK4 A0A0R1DY25 A0A0Q5VL17 A0A0Q5VZM5 A0A0Q5VLV6 B3MDI4 B4QIF3 A0A0J9U3T7 A0A0J9RFJ1 N6W6Y3 A0A0M4EVV7 A0A182JSX9 A0A0J9RE31 A0A0J9RFJ7 N6SZ40 A0A0J9U3U8 A0A0R3NQ16 A0A0R3NV63 A0A1A9UHE4 A0A0R3NQ74 A0A0R3NWD4 A0A0R3NQT2 A0A0R3NVL4 A0A1A9Z4G3 A0A1A9WWP0 X1WXH4 A0A1B0BZT0 B0WDG3 A0A1B0CG12 T1IJB9 B4H744 A0A2C9M9P4 A0A2C9M9S1 A0A2C9M9R5 A0A2C9M9R1 A0A2C9M9R2 A0A2C9MA25 A0A2C9M9R0 A0A2C9MA21 A0A2C9M9S0 K1RN38 A0A087URW9 A0A0V1MPD1 A0A0V1MPB9 A0A0V1H0V9 A0A0V1H0T9 A0A0V1EG07 A0A0V1IIL9 A0A0V1ABV8 A0A0V1CHM6 A0A0V0RHW1 A0A0V0RHP6 A0A0V0V2J5 A0A0V1BA73 A0A0V0RHH0 A0A0V0TF06 A0A0V1BAF1 A0A0V0TG64 A0A0V1BAH8 A0A0V1PG02 A0A0V1PFS2 A0A0V1H109 A0A0V1BA88
E1ZX90 A0A2J7Q5K2 F4WBT9 A0A139WDX9 A0A2A3EHV1 A0A195C3P2 A0A151WN78 A0A195FU85 A0A3L8E154 A0A158NW85 A0A195BD68 D6WVN9 A0A0L7QNY7 A0A195DI52 E2B9W7 A0A336LU78 A0A336LUI3 A0A0M8ZQ99 B4MIT1 A0A0R1DSR7 B3NP74 A0A0R1DUM0 A0A0R1DSJ0 A0A084WAX1 A1ZAJ2-2 A0A0R1DY12 A0A0Q5W0R5 U4U2Y2 A0A0Q5VLD5 A0A0R1DSK4 A0A0R1DY25 A0A0Q5VL17 A0A0Q5VZM5 A0A0Q5VLV6 B3MDI4 B4QIF3 A0A0J9U3T7 A0A0J9RFJ1 N6W6Y3 A0A0M4EVV7 A0A182JSX9 A0A0J9RE31 A0A0J9RFJ7 N6SZ40 A0A0J9U3U8 A0A0R3NQ16 A0A0R3NV63 A0A1A9UHE4 A0A0R3NQ74 A0A0R3NWD4 A0A0R3NQT2 A0A0R3NVL4 A0A1A9Z4G3 A0A1A9WWP0 X1WXH4 A0A1B0BZT0 B0WDG3 A0A1B0CG12 T1IJB9 B4H744 A0A2C9M9P4 A0A2C9M9S1 A0A2C9M9R5 A0A2C9M9R1 A0A2C9M9R2 A0A2C9MA25 A0A2C9M9R0 A0A2C9MA21 A0A2C9M9S0 K1RN38 A0A087URW9 A0A0V1MPD1 A0A0V1MPB9 A0A0V1H0V9 A0A0V1H0T9 A0A0V1EG07 A0A0V1IIL9 A0A0V1ABV8 A0A0V1CHM6 A0A0V0RHW1 A0A0V0RHP6 A0A0V0V2J5 A0A0V1BA73 A0A0V0RHH0 A0A0V0TF06 A0A0V1BAF1 A0A0V0TG64 A0A0V1BAH8 A0A0V1PG02 A0A0V1PFS2 A0A0V1H109 A0A0V1BA88
PDB
4UXS
E-value=1.44897e-136,
Score=1251
Ontologies
GO
GO:0008017
GO:0005524
GO:0003777
GO:0007018
GO:0005871
GO:0016192
GO:0016887
GO:0008574
GO:0005874
GO:0030705
GO:0003774
GO:0051963
GO:0047496
GO:0007528
GO:0046847
GO:0040012
GO:0032024
GO:0045886
GO:0016188
GO:0030742
GO:0008089
GO:0030424
GO:0030425
GO:0048490
GO:0048814
GO:0015031
GO:1904115
GO:0048489
GO:0045202
GO:0008345
GO:0007411
GO:0008088
GO:0005875
GO:0005515
GO:0019287
GO:0005634
GO:0006334
GO:0003779
GO:0016021
PANTHER
Topology
Subcellular location
Cytoplasm
Microtubule-associated. With evidence from 5 publications.
Cytoskeleton Microtubule-associated. With evidence from 5 publications.
Cell projection Microtubule-associated. With evidence from 5 publications.
Axon Microtubule-associated. With evidence from 5 publications.
Cytoskeleton Microtubule-associated. With evidence from 5 publications.
Cell projection Microtubule-associated. With evidence from 5 publications.
Axon Microtubule-associated. With evidence from 5 publications.
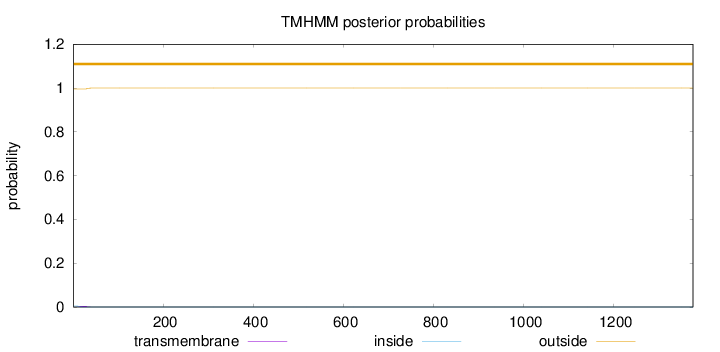
Length:
1376
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0824099999999999
Exp number, first 60 AAs:
0.08002
Total prob of N-in:
0.00430
outside
1 - 1376
Population Genetic Test Statistics
Pi
196.121142
Theta
172.163871
Tajima's D
0.712173
CLR
1.224044
CSRT
0.580020998950053
Interpretation
Uncertain
