Gene
KWMTBOMO01166
Pre Gene Modal
BGIBMGA007456
Annotation
PREDICTED:_kinesin-like_protein_unc-104_isoform_X4_[Bombyx_mori]
Full name
Kinesin-like protein unc-104
Alternative Name
Protein immaculate connections
Location in the cell
Mitochondrial Reliability : 2.269
Sequence
CDS
ATGACCCGCTTGGAGCAGGTGGGCCGCACGCGCCACCTCCTCGCACTCCGGGAGAGGCTGCGGCACGGACACGAGAACACGGTCGCGCCCAACGACTTCACCAAGACAGAGAAGGAGGTGTGCAACATGGCGGCGAAGGCGGCGGCCGAGGGCGGGCGCGACGAGCCCGTGTACGAGCCCTGGGACATGTCGCCCAAGGAACGAGAGCTCGCCACCAAATTCATTAAGCTCATGCAAGGAAGAATAGGTTCGACGGGCAAGGAAGTAGAGACCGCAGTGTCTCCCGCCACGCCCGTGGACGAGGGCGTTTCCGCTGACATCTCCACTCCGAGTCTCCTGTCGTCCATACACAGCGCTAGCAGCTTTGAGCTGTGCTCCCCGGAGCGCGCGATGCTGGAGAAGGCGGCGTGCGGGTGGGGGGCGGCGGGCGGCGCGCTGTACGTGCCCGACTGCGAGGAGGTGCGCGTGTCCGCCGCCGTGGCGCGCCGCGGACACCTCAACGTGCTGCAGCACGGCACGCACGGCTGGAAGAAACGCTGGCTGGTGGTCCGGAGGCCGTACGTGTTCATATACCGCGACGAGCGCGACCCCATCGAGCGCGCCGTCATCAACCTCGCCAACGCGCACGTCGAGTACTCCGAGGACCAGGAGCAGATGGTGCGCATGCCCAACACCTTCAGTGTGGTGAGCAAGGAGCGCGGCTACTTACTTCAGACGTTAAGCGACAAGGAGGTGCACGACTGGCTGTACGCGATCAACCCGCTGCTCGCCGGACAGATAAGGTCGCGGTCGGGCCGCCGCGGCGACAAGCCGGCGCCGCTGTCGGCATGA
Protein
MTRLEQVGRTRHLLALRERLRHGHENTVAPNDFTKTEKEVCNMAAKAAAEGGRDEPVYEPWDMSPKERELATKFIKLMQGRIGSTGKEVETAVSPATPVDEGVSADISTPSLLSSIHSASSFELCSPERAMLEKAACGWGAAGGALYVPDCEEVRVSAAVARRGHLNVLQHGTHGWKKRWLVVRRPYVFIYRDERDPIERAVINLANAHVEYSEDQEQMVRMPNTFSVVSKERGYLLQTLSDKEVHDWLYAINPLLAGQIRSRSGRRGDKPAPLSA
Summary
Description
Required for presynaptic maturation, has a role in axonal transport of dense-core vesicles carrying synaptic vesicle precursors, components required for the morphological transformation of axonal growth cones to mature boutons.
Subunit
Monomer (By similarity). Interacts with Arl8 (PubMed:30174114).
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. Unc-104 subfamily.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family. Unc-104 subfamily.
Keywords
Alternative splicing
ATP-binding
Cell projection
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Microtubule
Motor protein
Nucleotide-binding
Phosphoprotein
Protein transport
Reference proteome
Transport
Feature
chain Kinesin-like protein unc-104
splice variant In isoform E.
splice variant In isoform E.
Uniprot
H9JD59
A0A2A4JK74
A0A194QAA2
A0A2H1WUC8
A0A2A4JKZ3
A0A212EU86
+ More
A0A2W1BS23 A0A146LD53 A0A0K8T9H4 A0A0A9X329 A0A1B6M3I6 A0A0A9X260 A0A0C9RLS1 A0A0C9R567 K7IT34 A0A1B6GCQ3 A0A2S2NJT1 E1ZX90 A0A1B6JET4 A0A034V9U3 A0A1B6CAU0 A0A232FKZ6 A0A034V5H7 A0A026WLV1 A0A1B6CXY8 A0A1B6EGN6 B4H745 A0A2J7Q5K0 A0A2J7Q5K2 A0A2J7Q5K3 A0A069DYB2 A0A2J7Q5K1 A0A023F4N9 A0A2J7Q5K8 A0A088A1K2 A0A1B6ENW4 A0A0B4KF91 V9I8D8 A0A0Q5VLZ2 A0A0B4K767 A0A2M4B8N3 V9I998 A0A0R1DSQ4 A0A126GUN3 V9I754 A0A0Q5VL17 A0A0R1DY12 V9I8V6 A0A0J9U3U3 A0A0J9REJ2 A0A0J9RE31 A0A195C3P2 A0A0Q5VLV3 A0A0Q5VM46 A0A0Q5VU10 A0A0Q5VY14 A0A0B4K7D4 A1ZAJ2-3 A1ZAJ2 A0A0Q5VLD5 A0A0R1DWB8 A0A0Q5W0T5 A0A182GAH2 B4P5W2 A0A0Q5VLV6 A0A0R1DTA7 A0A0Q5W0R5 A1ZAJ2-2 A0A0R1DY40 B4HT85 A0A0R1DY25 A0A0R1DZ56 A0A0Q5VZM5 A0A0R1DUM0 B4QIF3 A0A0J9RFJ7 A0A0J9RFK3 A0A0J9RED2 A0A0J9RFJ1 A0A0J9REI7 B3NP74 A0A0J9U3T7 A0A0J9U3U8 X1WXH4 A0A0R1DSR7 A0A0R1DSJ0 A0A0R1DSK4 A0A0J9REC5 A0A0J9RE41 A0A1A9UHE4 A0A182P844 A0A2M4AF39 A0A1W4UY03 T1HYD7 A0A1W4UKR8 A0A1W4UXZ7 A0A182MYA1 A0A139WDX9 A0A2S2QWZ1 D6WVN9
A0A2W1BS23 A0A146LD53 A0A0K8T9H4 A0A0A9X329 A0A1B6M3I6 A0A0A9X260 A0A0C9RLS1 A0A0C9R567 K7IT34 A0A1B6GCQ3 A0A2S2NJT1 E1ZX90 A0A1B6JET4 A0A034V9U3 A0A1B6CAU0 A0A232FKZ6 A0A034V5H7 A0A026WLV1 A0A1B6CXY8 A0A1B6EGN6 B4H745 A0A2J7Q5K0 A0A2J7Q5K2 A0A2J7Q5K3 A0A069DYB2 A0A2J7Q5K1 A0A023F4N9 A0A2J7Q5K8 A0A088A1K2 A0A1B6ENW4 A0A0B4KF91 V9I8D8 A0A0Q5VLZ2 A0A0B4K767 A0A2M4B8N3 V9I998 A0A0R1DSQ4 A0A126GUN3 V9I754 A0A0Q5VL17 A0A0R1DY12 V9I8V6 A0A0J9U3U3 A0A0J9REJ2 A0A0J9RE31 A0A195C3P2 A0A0Q5VLV3 A0A0Q5VM46 A0A0Q5VU10 A0A0Q5VY14 A0A0B4K7D4 A1ZAJ2-3 A1ZAJ2 A0A0Q5VLD5 A0A0R1DWB8 A0A0Q5W0T5 A0A182GAH2 B4P5W2 A0A0Q5VLV6 A0A0R1DTA7 A0A0Q5W0R5 A1ZAJ2-2 A0A0R1DY40 B4HT85 A0A0R1DY25 A0A0R1DZ56 A0A0Q5VZM5 A0A0R1DUM0 B4QIF3 A0A0J9RFJ7 A0A0J9RFK3 A0A0J9RED2 A0A0J9RFJ1 A0A0J9REI7 B3NP74 A0A0J9U3T7 A0A0J9U3U8 X1WXH4 A0A0R1DSR7 A0A0R1DSJ0 A0A0R1DSK4 A0A0J9REC5 A0A0J9RE41 A0A1A9UHE4 A0A182P844 A0A2M4AF39 A0A1W4UY03 T1HYD7 A0A1W4UKR8 A0A1W4UXZ7 A0A182MYA1 A0A139WDX9 A0A2S2QWZ1 D6WVN9
Pubmed
EMBL
BABH01036281
BABH01036282
NWSH01001135
PCG72457.1
KQ459249
KPJ02359.1
+ More
ODYU01011089 SOQ56607.1 PCG72456.1 AGBW02012439 OWR45063.1 KZ149958 PZC76444.1 GDHC01012441 JAQ06188.1 GBRD01003619 JAG62202.1 GBHO01029538 GBHO01029536 JAG14066.1 JAG14068.1 GEBQ01009481 JAT30496.1 GBHO01029540 GBHO01029539 JAG14064.1 JAG14065.1 GBYB01007926 JAG77693.1 GBYB01007927 JAG77694.1 AAZX01006711 AAZX01008089 GECZ01009573 JAS60196.1 GGMR01004810 MBY17429.1 GL435030 EFN74180.1 GECU01010057 JAS97649.1 GAKP01020407 JAC38545.1 GEDC01026764 JAS10534.1 NNAY01000061 OXU31411.1 GAKP01020411 JAC38541.1 KK107154 EZA56943.1 GEDC01019020 JAS18278.1 GEDC01000210 JAS37088.1 CH479216 EDW33578.1 NEVH01017557 PNF23862.1 PNF23863.1 PNF23861.1 GBGD01000122 JAC88767.1 PNF23859.1 GBBI01002340 JAC16372.1 PNF23860.1 GECZ01030174 JAS39595.1 AE013599 AGB93565.1 JR036090 AEY56917.1 CH954179 KQS62419.1 AFH08126.1 GGFJ01000263 MBW49404.1 JR036084 AEY56914.1 CM000158 KRK00166.1 KRK00169.1 ALI30182.1 JR036086 AEY56915.1 KQS62413.1 KRK00162.1 JR036088 AEY56916.1 CM002911 KMY94345.1 KMY94346.1 KMY94338.1 KQ978317 KYM95220.1 KQS62422.1 KQS62417.1 KQS62421.1 KQS62414.1 AFH08127.1 AF247761 AAF74192.1 KQS62416.1 KRK00163.1 KRK00165.1 KQS62415.1 JXUM01050916 KQ561667 KXJ77862.1 EDW91878.1 KRK00164.1 KQS62423.1 KRK00175.1 KRK00178.1 KQS62424.1 KRK00170.1 CH480816 EDW48186.1 KRK00177.1 KRK00167.1 KQS62418.1 KQS62420.1 KRK00171.1 KRK00174.1 CM000362 EDX07400.1 KMY94336.1 KMY94342.1 KMY94343.1 KMY94344.1 KMY94349.1 KMY94347.1 KMY94339.1 KMY94341.1 EDV55713.1 KMY94340.1 KMY94350.1 ABLF02019510 KRK00173.1 KRK00176.1 KRK00168.1 KRK00172.1 KMY94337.1 KMY94348.1 GGFK01006062 MBW39383.1 ACPB03010694 ACPB03010695 ACPB03010696 ACPB03010697 KQ971357 KYB26178.1 GGMS01012980 MBY82183.1 EFA08595.1
ODYU01011089 SOQ56607.1 PCG72456.1 AGBW02012439 OWR45063.1 KZ149958 PZC76444.1 GDHC01012441 JAQ06188.1 GBRD01003619 JAG62202.1 GBHO01029538 GBHO01029536 JAG14066.1 JAG14068.1 GEBQ01009481 JAT30496.1 GBHO01029540 GBHO01029539 JAG14064.1 JAG14065.1 GBYB01007926 JAG77693.1 GBYB01007927 JAG77694.1 AAZX01006711 AAZX01008089 GECZ01009573 JAS60196.1 GGMR01004810 MBY17429.1 GL435030 EFN74180.1 GECU01010057 JAS97649.1 GAKP01020407 JAC38545.1 GEDC01026764 JAS10534.1 NNAY01000061 OXU31411.1 GAKP01020411 JAC38541.1 KK107154 EZA56943.1 GEDC01019020 JAS18278.1 GEDC01000210 JAS37088.1 CH479216 EDW33578.1 NEVH01017557 PNF23862.1 PNF23863.1 PNF23861.1 GBGD01000122 JAC88767.1 PNF23859.1 GBBI01002340 JAC16372.1 PNF23860.1 GECZ01030174 JAS39595.1 AE013599 AGB93565.1 JR036090 AEY56917.1 CH954179 KQS62419.1 AFH08126.1 GGFJ01000263 MBW49404.1 JR036084 AEY56914.1 CM000158 KRK00166.1 KRK00169.1 ALI30182.1 JR036086 AEY56915.1 KQS62413.1 KRK00162.1 JR036088 AEY56916.1 CM002911 KMY94345.1 KMY94346.1 KMY94338.1 KQ978317 KYM95220.1 KQS62422.1 KQS62417.1 KQS62421.1 KQS62414.1 AFH08127.1 AF247761 AAF74192.1 KQS62416.1 KRK00163.1 KRK00165.1 KQS62415.1 JXUM01050916 KQ561667 KXJ77862.1 EDW91878.1 KRK00164.1 KQS62423.1 KRK00175.1 KRK00178.1 KQS62424.1 KRK00170.1 CH480816 EDW48186.1 KRK00177.1 KRK00167.1 KQS62418.1 KQS62420.1 KRK00171.1 KRK00174.1 CM000362 EDX07400.1 KMY94336.1 KMY94342.1 KMY94343.1 KMY94344.1 KMY94349.1 KMY94347.1 KMY94339.1 KMY94341.1 EDV55713.1 KMY94340.1 KMY94350.1 ABLF02019510 KRK00173.1 KRK00176.1 KRK00168.1 KRK00172.1 KMY94337.1 KMY94348.1 GGFK01006062 MBW39383.1 ACPB03010694 ACPB03010695 ACPB03010696 ACPB03010697 KQ971357 KYB26178.1 GGMS01012980 MBY82183.1 EFA08595.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000002358
UP000000311
+ More
UP000215335 UP000053097 UP000008744 UP000235965 UP000005203 UP000000803 UP000008711 UP000002282 UP000078542 UP000069940 UP000249989 UP000001292 UP000000304 UP000007819 UP000078200 UP000075885 UP000192221 UP000015103 UP000075884 UP000007266
UP000215335 UP000053097 UP000008744 UP000235965 UP000005203 UP000000803 UP000008711 UP000002282 UP000078542 UP000069940 UP000249989 UP000001292 UP000000304 UP000007819 UP000078200 UP000075885 UP000192221 UP000015103 UP000075884 UP000007266
Pfam
Interpro
IPR011993
PH-like_dom_sf
+ More
IPR001849 PH_domain
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR032405 Kinesin_assoc
IPR036961 Kinesin_motor_dom_sf
IPR001752 Kinesin_motor_dom
IPR022140 Kinesin-like_KIF1-typ
IPR027640 Kinesin-like_fam
IPR008984 SMAD_FHA_dom_sf
IPR000253 FHA_dom
IPR022164 Kinesin-like
IPR004015 SKI-int_prot_SKIP_SNW-dom
IPR001849 PH_domain
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR032405 Kinesin_assoc
IPR036961 Kinesin_motor_dom_sf
IPR001752 Kinesin_motor_dom
IPR022140 Kinesin-like_KIF1-typ
IPR027640 Kinesin-like_fam
IPR008984 SMAD_FHA_dom_sf
IPR000253 FHA_dom
IPR022164 Kinesin-like
IPR004015 SKI-int_prot_SKIP_SNW-dom
Gene 3D
CDD
ProteinModelPortal
H9JD59
A0A2A4JK74
A0A194QAA2
A0A2H1WUC8
A0A2A4JKZ3
A0A212EU86
+ More
A0A2W1BS23 A0A146LD53 A0A0K8T9H4 A0A0A9X329 A0A1B6M3I6 A0A0A9X260 A0A0C9RLS1 A0A0C9R567 K7IT34 A0A1B6GCQ3 A0A2S2NJT1 E1ZX90 A0A1B6JET4 A0A034V9U3 A0A1B6CAU0 A0A232FKZ6 A0A034V5H7 A0A026WLV1 A0A1B6CXY8 A0A1B6EGN6 B4H745 A0A2J7Q5K0 A0A2J7Q5K2 A0A2J7Q5K3 A0A069DYB2 A0A2J7Q5K1 A0A023F4N9 A0A2J7Q5K8 A0A088A1K2 A0A1B6ENW4 A0A0B4KF91 V9I8D8 A0A0Q5VLZ2 A0A0B4K767 A0A2M4B8N3 V9I998 A0A0R1DSQ4 A0A126GUN3 V9I754 A0A0Q5VL17 A0A0R1DY12 V9I8V6 A0A0J9U3U3 A0A0J9REJ2 A0A0J9RE31 A0A195C3P2 A0A0Q5VLV3 A0A0Q5VM46 A0A0Q5VU10 A0A0Q5VY14 A0A0B4K7D4 A1ZAJ2-3 A1ZAJ2 A0A0Q5VLD5 A0A0R1DWB8 A0A0Q5W0T5 A0A182GAH2 B4P5W2 A0A0Q5VLV6 A0A0R1DTA7 A0A0Q5W0R5 A1ZAJ2-2 A0A0R1DY40 B4HT85 A0A0R1DY25 A0A0R1DZ56 A0A0Q5VZM5 A0A0R1DUM0 B4QIF3 A0A0J9RFJ7 A0A0J9RFK3 A0A0J9RED2 A0A0J9RFJ1 A0A0J9REI7 B3NP74 A0A0J9U3T7 A0A0J9U3U8 X1WXH4 A0A0R1DSR7 A0A0R1DSJ0 A0A0R1DSK4 A0A0J9REC5 A0A0J9RE41 A0A1A9UHE4 A0A182P844 A0A2M4AF39 A0A1W4UY03 T1HYD7 A0A1W4UKR8 A0A1W4UXZ7 A0A182MYA1 A0A139WDX9 A0A2S2QWZ1 D6WVN9
A0A2W1BS23 A0A146LD53 A0A0K8T9H4 A0A0A9X329 A0A1B6M3I6 A0A0A9X260 A0A0C9RLS1 A0A0C9R567 K7IT34 A0A1B6GCQ3 A0A2S2NJT1 E1ZX90 A0A1B6JET4 A0A034V9U3 A0A1B6CAU0 A0A232FKZ6 A0A034V5H7 A0A026WLV1 A0A1B6CXY8 A0A1B6EGN6 B4H745 A0A2J7Q5K0 A0A2J7Q5K2 A0A2J7Q5K3 A0A069DYB2 A0A2J7Q5K1 A0A023F4N9 A0A2J7Q5K8 A0A088A1K2 A0A1B6ENW4 A0A0B4KF91 V9I8D8 A0A0Q5VLZ2 A0A0B4K767 A0A2M4B8N3 V9I998 A0A0R1DSQ4 A0A126GUN3 V9I754 A0A0Q5VL17 A0A0R1DY12 V9I8V6 A0A0J9U3U3 A0A0J9REJ2 A0A0J9RE31 A0A195C3P2 A0A0Q5VLV3 A0A0Q5VM46 A0A0Q5VU10 A0A0Q5VY14 A0A0B4K7D4 A1ZAJ2-3 A1ZAJ2 A0A0Q5VLD5 A0A0R1DWB8 A0A0Q5W0T5 A0A182GAH2 B4P5W2 A0A0Q5VLV6 A0A0R1DTA7 A0A0Q5W0R5 A1ZAJ2-2 A0A0R1DY40 B4HT85 A0A0R1DY25 A0A0R1DZ56 A0A0Q5VZM5 A0A0R1DUM0 B4QIF3 A0A0J9RFJ7 A0A0J9RFK3 A0A0J9RED2 A0A0J9RFJ1 A0A0J9REI7 B3NP74 A0A0J9U3T7 A0A0J9U3U8 X1WXH4 A0A0R1DSR7 A0A0R1DSJ0 A0A0R1DSK4 A0A0J9REC5 A0A0J9RE41 A0A1A9UHE4 A0A182P844 A0A2M4AF39 A0A1W4UY03 T1HYD7 A0A1W4UKR8 A0A1W4UXZ7 A0A182MYA1 A0A139WDX9 A0A2S2QWZ1 D6WVN9
PDB
1EAZ
E-value=0.0170618,
Score=88
Ontologies
GO
GO:0007018
GO:0008017
GO:0005524
GO:0003777
GO:0005681
GO:0000398
GO:0016887
GO:0016192
GO:0051963
GO:0030742
GO:0016188
GO:0045886
GO:0032024
GO:0046847
GO:0040012
GO:0007528
GO:0047496
GO:0015031
GO:0005874
GO:0048814
GO:0048490
GO:0030425
GO:0030424
GO:0008089
GO:0005875
GO:0008088
GO:0007411
GO:0045202
GO:0008345
GO:0048489
GO:1904115
GO:0016021
GO:0005871
GO:0008574
GO:0030705
PANTHER
Topology
Subcellular location
Cytoplasm
Microtubule-associated. With evidence from 5 publications.
Cytoskeleton Microtubule-associated. With evidence from 5 publications.
Cell projection Microtubule-associated. With evidence from 5 publications.
Axon Microtubule-associated. With evidence from 5 publications.
Cytoskeleton Microtubule-associated. With evidence from 5 publications.
Cell projection Microtubule-associated. With evidence from 5 publications.
Axon Microtubule-associated. With evidence from 5 publications.
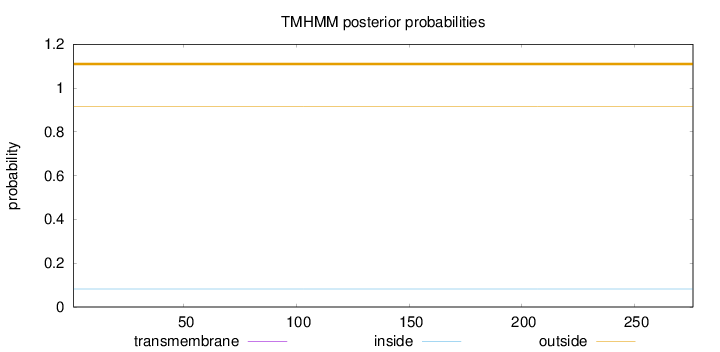
Length:
276
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000720000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08276
outside
1 - 276
Population Genetic Test Statistics
Pi
249.226165
Theta
18.567629
Tajima's D
-1.304764
CLR
0.890088
CSRT
0.0858957052147393
Interpretation
Uncertain
