Pre Gene Modal
BGIBMGA007486
Annotation
PREDICTED:_vacuolar_protein_sorting-associated_protein_11_homolog_[Bombyx_mori]
Full name
Vacuolar protein sorting-associated protein 11 homolog
Location in the cell
Nuclear Reliability : 2.505
Sequence
CDS
ATGCTACATCCATTCTCTTGGTGCTTCGCCGGTACAGTCGACAAGAACTACGAAAGTTACTCCGTAGTATCAGAGTACTACGGACGCGGTCTGTTCAACAAGCTGACCGTCGTTACGGAGAGCGAGGAAGCCGCCCCCGCGCCCGCCTCCGCGCCCGTCCCCGCACCCGTCCCCGCACCCGCGCCCGCCCCCGCCCCCGCCTATGGTGCCGGAGCTGAAGCTAAACTCAGGATGCAGGAAGGACAAAGCAAGCAGTTGTTCGTACCGAACGCTCCGAAACAGATAGCCCCTAAAGGCAGCAGCGTGGTGCCGGTCCCCGAGGGCAGGATGCGCCTCATGGAGCAGCAGCAGTACAGCAGCAGCCTCGAGGCGAACCTCACCAGGCTCGAGCCCGTGCACACCAAGAAACCGTCGCCCTTCGCCTCGCCGAGGGCCGCGAAGCGAATAGTCCTCGACCAACCTAAAACCTCGAAACCTCCAGTAACGAAGCTCGAGGGGAGCAACCCCTTCGACGAGCACTACGACGAGTCCAAGAACCCGTTCGCGGACGAACAAGGAGACGACCCGACCAACCCCTTCGCGGGAGACGACGACTACGACTCCAACTATAATCCCTTCGCTAAAGACGACTAG
Protein
MLHPFSWCFAGTVDKNYESYSVVSEYYGRGLFNKLTVVTESEEAAPAPASAPVPAPVPAPAPAPAPAYGAGAEAKLRMQEGQSKQLFVPNAPKQIAPKGSSVVPVPEGRMRLMEQQQYSSSLEANLTRLEPVHTKKPSPFASPRAAKRIVLDQPKTSKPPVTKLEGSNPFDEHYDESKNPFADEQGDDPTNPFAGDDDYDSNYNPFAKDD
Summary
Similarity
Belongs to the VPS11 family.
Uniprot
EMBL
BABH01036280
GDQN01011132
JAT79922.1
GDQN01008598
JAT82456.1
ODYU01012745
+ More
SOQ59209.1 KZ149958 PZC76445.1 KQ459249 KPJ02358.1 KQ460366 KPJ15599.1 NEVH01026389 PNF14340.1 PNF14339.1 PNF14338.1 GFDF01003494 JAV10590.1 GL445407 EFN89642.1 KQ982431 KYQ56510.1 GBYB01009735 JAG79502.1 ADTU01002180 ADTU01002181 KQ435711 KOX79645.1 KQ414631 KOC67277.1 LBMM01001510 KMQ96170.1
SOQ59209.1 KZ149958 PZC76445.1 KQ459249 KPJ02358.1 KQ460366 KPJ15599.1 NEVH01026389 PNF14340.1 PNF14339.1 PNF14338.1 GFDF01003494 JAV10590.1 GL445407 EFN89642.1 KQ982431 KYQ56510.1 GBYB01009735 JAG79502.1 ADTU01002180 ADTU01002181 KQ435711 KOX79645.1 KQ414631 KOC67277.1 LBMM01001510 KMQ96170.1
Proteomes
PRIDE
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR001841 Znf_RING
IPR011990 TPR-like_helical_dom_sf
IPR016528 VPS11
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR000547 Clathrin_H-chain/VPS_repeat
IPR013083 Znf_RING/FYVE/PHD
IPR027791 Galactosyl_T_C
IPR003859 Galactosyl_T
IPR027995 Galactosyl_T_N
IPR029044 Nucleotide-diphossugar_trans
IPR024763 VPS11_C
IPR016024 ARM-type_fold
IPR001680 WD40_repeat
IPR001841 Znf_RING
IPR011990 TPR-like_helical_dom_sf
IPR016528 VPS11
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR000547 Clathrin_H-chain/VPS_repeat
IPR013083 Znf_RING/FYVE/PHD
IPR027791 Galactosyl_T_C
IPR003859 Galactosyl_T
IPR027995 Galactosyl_T_N
IPR029044 Nucleotide-diphossugar_trans
IPR024763 VPS11_C
IPR016024 ARM-type_fold
IPR001680 WD40_repeat
Gene 3D
ProteinModelPortal
Ontologies
GO
PANTHER
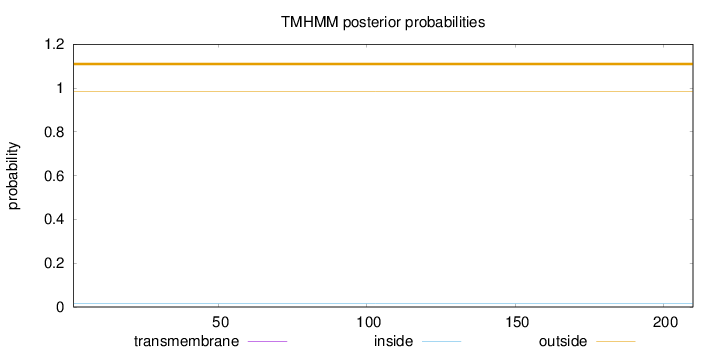
Topology
Length:
210
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00022
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.01437
outside
1 - 210
Population Genetic Test Statistics
Pi
166.037725
Theta
125.108512
Tajima's D
0.568256
CLR
2.621354
CSRT
0.533423328833558
Interpretation
Uncertain
