Gene
KWMTBOMO01160
Annotation
PREDICTED:_RNA-directed_DNA_polymerase_from_mobile_element_jockey-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.638
Sequence
CDS
ATGATCCTCCTGCTTGTGGATGAGCTTATCCCTATCATTACATATATTCTTAACTTCTCTCTCCTAAAAGGCTCATTCCCAAGTAGCTGGAGAAAGGCTCATGTTCTCCCACTTCCTAAGGTCCCTAATCCTTCCTCTTTTTCTCAGTTTCGTCCAATCTCAATACTTCCCATTCTCTCTAAAGTCTTGGAGGGTTTAGTCCAAAGACAGCTCACTGTCTTTCTTAATAAAAACAATCTCCTCAACTCTTTTCAGTCTGGTTTCCGCCAAGGCCACAGCACGGTCACTGCATTGCTCAAGGTCACCGACGACATTCGCCACGCGATTGATGTGGGGCAGCTAACCGTGTTGACCTTATTGGACTTCAGCAGTGCATTTAACTCAGTAGACTTTGACATCCTTCTAAGTATCCTTGGATCTCTTAATGTATCTCCACTTGCTCTAACCTGGTTTGAATCTTATCTTCGGGGACGCTCGCAATGTGTCCGCTCTGATGATGCTACATCGAAATGGTGTGATTTAACTGTGGGTGTACCTCAAGGTGGCGTACTATCTCCTCTGCTTTTCTCTATTTTTATTAACGAAATCACCCAGGTGCTGACTTCTCACTACCATCTCTACGCTGACGATTTGCAGATTTATAGACATTTTAAAGCTGATGATGTTGCTCTAGCTATTGCTGAAATCAATGGAGATCTTGATAGAATTAGTGCTTGGGCTAAATCTTTTGGACTTCTTGTGAATCCATCAAAATCACAGGTCATAGTAATTGGTAGTTCCTATAGGTACAACACGATTGACCACATTAATTTGCCCCCTGTGACGCTAGATGGAATTCCAATCGCTTATTCTAAAACAGCAAAAAACCTAGGTATCGTTCTTGATTGTCATCTCTCTTGGTCCAGTCAGATAAATTTAGTTAGCAGGAAAGTGCATTGCACGTTTCAGTCTCTCAAGCGTCTTCAGCGTTTCTTGCCCTTTGAAGCTAAAGTCACCCTGGCCCAATCCCTTATGCTTCCTCTCCTTGACTATGCGGATGTCTGTTTCCTCGACGCCGCGGAGGGGCTATGGAACAAACTGGAAAGGTTACAAAATTTATGCATACGCTTTATTTTTGGTTTACGCAAGTATGATCATGTCTCGCAGTACAGATCACAACTTAATTGGTTGCCTATCCGACTTCGCCGTGATGCACATGTTTTATGTATGCTGTTCAACATCCTCAATAACCCAGCTGTTCCCCAGTACCTCCGTGAGCGCTTCGAGTACCAGATGCCCCGTGGTGTACCCTGTCGTACAAGTAGATCCCTTCTCCTTAATTTTCCCCCTCACTCCTCCAGCCGCTACGGTGGTTCGTTCACTGTGCACTCTGTGCGTCTGTGGAACTCAGTTCCACACTCAATCCGTGCAAGTCCCTCACTTGAAGTTTTCAGGAAGGCATTAAAGGAGCACTATCTATCAATTTAG
Protein
MILLLVDELIPIITYILNFSLLKGSFPSSWRKAHVLPLPKVPNPSSFSQFRPISILPILSKVLEGLVQRQLTVFLNKNNLLNSFQSGFRQGHSTVTALLKVTDDIRHAIDVGQLTVLTLLDFSSAFNSVDFDILLSILGSLNVSPLALTWFESYLRGRSQCVRSDDATSKWCDLTVGVPQGGVLSPLLFSIFINEITQVLTSHYHLYADDLQIYRHFKADDVALAIAEINGDLDRISAWAKSFGLLVNPSKSQVIVIGSSYRYNTIDHINLPPVTLDGIPIAYSKTAKNLGIVLDCHLSWSSQINLVSRKVHCTFQSLKRLQRFLPFEAKVTLAQSLMLPLLDYADVCFLDAAEGLWNKLERLQNLCIRFIFGLRKYDHVSQYRSQLNWLPIRLRRDAHVLCMLFNILNNPAVPQYLRERFEYQMPRGVPCRTSRSLLLNFPPHSSSRYGGSFTVHSVRLWNSVPHSIRASPSLEVFRKALKEHYLSI
Summary
Uniprot
A0A2A4JRM8
A0A2A4JWP8
A0A1B6L226
A0A1B6L6Y3
A0A1B6KKS7
A0A1B6LTE5
+ More
A0A1B6K9B1 A0A2H1WCC1 A0A1B6HKJ1 U5EQQ2 A0A023EXE1 U5EUG4 U5EQA6 A0A194QKM3 A0A0J7N335 A0A0A9VTC1 A0A354GGU9 A0A1B6K7R5 A0A354GGZ3 K7JAP0 A0A0J7K589 A0A0J7KR96 A0A0J7KSZ7 A0A354GIY0 A0A3L8DMY5 A0A3C1S155 A0A354GES4 A0A3C1RY27 A0A3C1S1N3 A0A3C1S319 A0A146MD70 A0A0A9WX05 A0A0A9YXJ4 A0A354GG23 A0A3C1S3J7 A0A3C1RZS6 A0A1Y1MCW4 A0A0A9YIX2 A0A354GI57 U5ETW8 A0A354GHC6 A0A1Y1MMA0 A0A3C1S2P1 A0A2A4IWQ4 A0A2A4KAV4 U5EQT8 A0A3P8SQQ3 A0A1Y1M988 A0A1Y1NB24 A0A2W1BK42 U5EQV9 A0A0A9WM49 U5ETR7 A0A194QGG5 U5EQJ2 M3XKU2 A0A3P9MM16 A0A1L8DJB0 A0A0J7KF45 A0A3B1J4H0 U5EQE2 U5ENG8 K7JTU8 A0A1Y1M3U5 A0A3B1ID12 A0A0A9Y046 A0A1B6KNH6 A0A2B4RHP5 A0A2G8LDL3 A0A3P9H3I2 A0A3B1IFS8 A0A1Y1MBG6 A0A3P9M6K8 U5EEC7 A0A3P9IQD1 A0A3P8NL25 W8AIM4 R4GCU3 A0A3B3C4H2 R4GD80 W8B6W6 A0A1Y1LVZ3 A0A3B3BIG0 U5ER05 A0A1A8PRG6 A0A3B1ITJ3 U5EQB9 A0A3P8P827 A0A3P8P7I2 A0A1A8R6J3 U5EN99 K7JMB3 A0A3B3B5V6 A0A3B3BWV2 R4G973 A0A3B3DGF3 A0A3P8PXB5 A0A3B1IVJ9
A0A1B6K9B1 A0A2H1WCC1 A0A1B6HKJ1 U5EQQ2 A0A023EXE1 U5EUG4 U5EQA6 A0A194QKM3 A0A0J7N335 A0A0A9VTC1 A0A354GGU9 A0A1B6K7R5 A0A354GGZ3 K7JAP0 A0A0J7K589 A0A0J7KR96 A0A0J7KSZ7 A0A354GIY0 A0A3L8DMY5 A0A3C1S155 A0A354GES4 A0A3C1RY27 A0A3C1S1N3 A0A3C1S319 A0A146MD70 A0A0A9WX05 A0A0A9YXJ4 A0A354GG23 A0A3C1S3J7 A0A3C1RZS6 A0A1Y1MCW4 A0A0A9YIX2 A0A354GI57 U5ETW8 A0A354GHC6 A0A1Y1MMA0 A0A3C1S2P1 A0A2A4IWQ4 A0A2A4KAV4 U5EQT8 A0A3P8SQQ3 A0A1Y1M988 A0A1Y1NB24 A0A2W1BK42 U5EQV9 A0A0A9WM49 U5ETR7 A0A194QGG5 U5EQJ2 M3XKU2 A0A3P9MM16 A0A1L8DJB0 A0A0J7KF45 A0A3B1J4H0 U5EQE2 U5ENG8 K7JTU8 A0A1Y1M3U5 A0A3B1ID12 A0A0A9Y046 A0A1B6KNH6 A0A2B4RHP5 A0A2G8LDL3 A0A3P9H3I2 A0A3B1IFS8 A0A1Y1MBG6 A0A3P9M6K8 U5EEC7 A0A3P9IQD1 A0A3P8NL25 W8AIM4 R4GCU3 A0A3B3C4H2 R4GD80 W8B6W6 A0A1Y1LVZ3 A0A3B3BIG0 U5ER05 A0A1A8PRG6 A0A3B1ITJ3 U5EQB9 A0A3P8P827 A0A3P8P7I2 A0A1A8R6J3 U5EN99 K7JMB3 A0A3B3B5V6 A0A3B3BWV2 R4G973 A0A3B3DGF3 A0A3P8PXB5 A0A3B1IVJ9
Pubmed
EMBL
NWSH01000817
PCG74070.1
NWSH01000468
PCG76206.1
GEBQ01022205
JAT17772.1
+ More
GEBQ01020572 JAT19405.1 GEBQ01027915 JAT12062.1 GEBQ01013060 GEBQ01010336 JAT26917.1 JAT29641.1 GEBQ01031972 JAT08005.1 ODYU01007705 SOQ50729.1 GECU01032510 JAS75196.1 GANO01004126 JAB55745.1 GBBI01004799 JAC13913.1 GANO01003893 JAB55978.1 GANO01004338 JAB55533.1 KQ461198 KPJ06072.1 LBMM01010997 KMQ87090.1 GBHO01045128 JAF98475.1 DNUT01000248 HBI40237.1 GECU01000221 JAT07486.1 DNUT01000258 HBI40281.1 AAZX01023386 LBMM01014048 KMQ85356.1 LBMM01004152 KMQ92759.1 LBMM01003452 KMQ93522.1 DNUT01000451 HBI40968.1 QOIP01000006 RLU21552.1 DMQF01000113 HAO14334.1 DNUT01000056 HBI39512.1 DMQF01000003 HAO13943.1 DMQF01000352 HAO15199.1 DMQF01000396 HAO15350.1 GDHC01000945 JAQ17684.1 GBHO01031326 GBHO01031325 JAG12278.1 JAG12279.1 GBHO01007781 JAG35823.1 DNUT01000170 HBI39961.1 DMQF01000436 HAO15547.1 DMQF01000157 HAO14486.1 GEZM01034710 JAV83652.1 GBHO01012041 GBHO01012038 JAG31563.1 JAG31566.1 DNUT01000374 HBI40695.1 GANO01004166 JAB55705.1 DNUT01000297 HBI40414.1 GEZM01027174 GEZM01027173 JAV86813.1 DMQF01000361 HAO15230.1 NWSH01006271 PCG63543.1 NWSH01000010 PCG80923.1 GANO01004070 JAB55801.1 GEZM01039829 JAV81150.1 GEZM01007604 JAV95063.1 KZ150070 PZC74075.1 GANO01004066 JAB55805.1 GBHO01034750 JAG08854.1 GANO01004238 JAB55633.1 KQ458880 KPJ04628.1 GANO01004258 JAB55613.1 AFYH01007667 GFDF01007660 JAV06424.1 LBMM01008498 KMQ88826.1 GANO01004325 JAB55546.1 GANO01004094 JAB55777.1 AAZX01002422 AAZX01014809 GEZM01041318 GEZM01041317 JAV80524.1 GBHO01019121 JAG24483.1 GEBQ01026990 JAT12987.1 LSMT01000554 PFX16333.1 MRZV01000114 PIK58376.1 GEZM01037808 JAV81920.1 GANO01004362 JAB55509.1 GAMC01021038 GAMC01021033 GAMC01021031 JAB85522.1 GAMC01021037 GAMC01021036 JAB85519.1 GEZM01045590 GEZM01045589 JAV77734.1 GANO01003995 JAB55876.1 HAEI01003198 SBR83602.1 GANO01004361 JAB55510.1 HAEG01016041 SBS01700.1 GANO01004198 JAB55673.1 AAZX01023421 AAZX01025062 AAWZ02022173
GEBQ01020572 JAT19405.1 GEBQ01027915 JAT12062.1 GEBQ01013060 GEBQ01010336 JAT26917.1 JAT29641.1 GEBQ01031972 JAT08005.1 ODYU01007705 SOQ50729.1 GECU01032510 JAS75196.1 GANO01004126 JAB55745.1 GBBI01004799 JAC13913.1 GANO01003893 JAB55978.1 GANO01004338 JAB55533.1 KQ461198 KPJ06072.1 LBMM01010997 KMQ87090.1 GBHO01045128 JAF98475.1 DNUT01000248 HBI40237.1 GECU01000221 JAT07486.1 DNUT01000258 HBI40281.1 AAZX01023386 LBMM01014048 KMQ85356.1 LBMM01004152 KMQ92759.1 LBMM01003452 KMQ93522.1 DNUT01000451 HBI40968.1 QOIP01000006 RLU21552.1 DMQF01000113 HAO14334.1 DNUT01000056 HBI39512.1 DMQF01000003 HAO13943.1 DMQF01000352 HAO15199.1 DMQF01000396 HAO15350.1 GDHC01000945 JAQ17684.1 GBHO01031326 GBHO01031325 JAG12278.1 JAG12279.1 GBHO01007781 JAG35823.1 DNUT01000170 HBI39961.1 DMQF01000436 HAO15547.1 DMQF01000157 HAO14486.1 GEZM01034710 JAV83652.1 GBHO01012041 GBHO01012038 JAG31563.1 JAG31566.1 DNUT01000374 HBI40695.1 GANO01004166 JAB55705.1 DNUT01000297 HBI40414.1 GEZM01027174 GEZM01027173 JAV86813.1 DMQF01000361 HAO15230.1 NWSH01006271 PCG63543.1 NWSH01000010 PCG80923.1 GANO01004070 JAB55801.1 GEZM01039829 JAV81150.1 GEZM01007604 JAV95063.1 KZ150070 PZC74075.1 GANO01004066 JAB55805.1 GBHO01034750 JAG08854.1 GANO01004238 JAB55633.1 KQ458880 KPJ04628.1 GANO01004258 JAB55613.1 AFYH01007667 GFDF01007660 JAV06424.1 LBMM01008498 KMQ88826.1 GANO01004325 JAB55546.1 GANO01004094 JAB55777.1 AAZX01002422 AAZX01014809 GEZM01041318 GEZM01041317 JAV80524.1 GBHO01019121 JAG24483.1 GEBQ01026990 JAT12987.1 LSMT01000554 PFX16333.1 MRZV01000114 PIK58376.1 GEZM01037808 JAV81920.1 GANO01004362 JAB55509.1 GAMC01021038 GAMC01021033 GAMC01021031 JAB85522.1 GAMC01021037 GAMC01021036 JAB85519.1 GEZM01045590 GEZM01045589 JAV77734.1 GANO01003995 JAB55876.1 HAEI01003198 SBR83602.1 GANO01004361 JAB55510.1 HAEG01016041 SBS01700.1 GANO01004198 JAB55673.1 AAZX01023421 AAZX01025062 AAWZ02022173
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JRM8
A0A2A4JWP8
A0A1B6L226
A0A1B6L6Y3
A0A1B6KKS7
A0A1B6LTE5
+ More
A0A1B6K9B1 A0A2H1WCC1 A0A1B6HKJ1 U5EQQ2 A0A023EXE1 U5EUG4 U5EQA6 A0A194QKM3 A0A0J7N335 A0A0A9VTC1 A0A354GGU9 A0A1B6K7R5 A0A354GGZ3 K7JAP0 A0A0J7K589 A0A0J7KR96 A0A0J7KSZ7 A0A354GIY0 A0A3L8DMY5 A0A3C1S155 A0A354GES4 A0A3C1RY27 A0A3C1S1N3 A0A3C1S319 A0A146MD70 A0A0A9WX05 A0A0A9YXJ4 A0A354GG23 A0A3C1S3J7 A0A3C1RZS6 A0A1Y1MCW4 A0A0A9YIX2 A0A354GI57 U5ETW8 A0A354GHC6 A0A1Y1MMA0 A0A3C1S2P1 A0A2A4IWQ4 A0A2A4KAV4 U5EQT8 A0A3P8SQQ3 A0A1Y1M988 A0A1Y1NB24 A0A2W1BK42 U5EQV9 A0A0A9WM49 U5ETR7 A0A194QGG5 U5EQJ2 M3XKU2 A0A3P9MM16 A0A1L8DJB0 A0A0J7KF45 A0A3B1J4H0 U5EQE2 U5ENG8 K7JTU8 A0A1Y1M3U5 A0A3B1ID12 A0A0A9Y046 A0A1B6KNH6 A0A2B4RHP5 A0A2G8LDL3 A0A3P9H3I2 A0A3B1IFS8 A0A1Y1MBG6 A0A3P9M6K8 U5EEC7 A0A3P9IQD1 A0A3P8NL25 W8AIM4 R4GCU3 A0A3B3C4H2 R4GD80 W8B6W6 A0A1Y1LVZ3 A0A3B3BIG0 U5ER05 A0A1A8PRG6 A0A3B1ITJ3 U5EQB9 A0A3P8P827 A0A3P8P7I2 A0A1A8R6J3 U5EN99 K7JMB3 A0A3B3B5V6 A0A3B3BWV2 R4G973 A0A3B3DGF3 A0A3P8PXB5 A0A3B1IVJ9
A0A1B6K9B1 A0A2H1WCC1 A0A1B6HKJ1 U5EQQ2 A0A023EXE1 U5EUG4 U5EQA6 A0A194QKM3 A0A0J7N335 A0A0A9VTC1 A0A354GGU9 A0A1B6K7R5 A0A354GGZ3 K7JAP0 A0A0J7K589 A0A0J7KR96 A0A0J7KSZ7 A0A354GIY0 A0A3L8DMY5 A0A3C1S155 A0A354GES4 A0A3C1RY27 A0A3C1S1N3 A0A3C1S319 A0A146MD70 A0A0A9WX05 A0A0A9YXJ4 A0A354GG23 A0A3C1S3J7 A0A3C1RZS6 A0A1Y1MCW4 A0A0A9YIX2 A0A354GI57 U5ETW8 A0A354GHC6 A0A1Y1MMA0 A0A3C1S2P1 A0A2A4IWQ4 A0A2A4KAV4 U5EQT8 A0A3P8SQQ3 A0A1Y1M988 A0A1Y1NB24 A0A2W1BK42 U5EQV9 A0A0A9WM49 U5ETR7 A0A194QGG5 U5EQJ2 M3XKU2 A0A3P9MM16 A0A1L8DJB0 A0A0J7KF45 A0A3B1J4H0 U5EQE2 U5ENG8 K7JTU8 A0A1Y1M3U5 A0A3B1ID12 A0A0A9Y046 A0A1B6KNH6 A0A2B4RHP5 A0A2G8LDL3 A0A3P9H3I2 A0A3B1IFS8 A0A1Y1MBG6 A0A3P9M6K8 U5EEC7 A0A3P9IQD1 A0A3P8NL25 W8AIM4 R4GCU3 A0A3B3C4H2 R4GD80 W8B6W6 A0A1Y1LVZ3 A0A3B3BIG0 U5ER05 A0A1A8PRG6 A0A3B1ITJ3 U5EQB9 A0A3P8P827 A0A3P8P7I2 A0A1A8R6J3 U5EN99 K7JMB3 A0A3B3B5V6 A0A3B3BWV2 R4G973 A0A3B3DGF3 A0A3P8PXB5 A0A3B1IVJ9
PDB
6AR3
E-value=1.02142e-05,
Score=118
Ontologies
KEGG
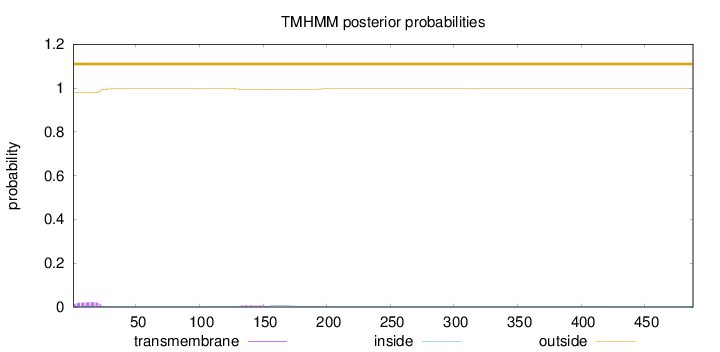
Topology
Length:
488
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.648799999999999
Exp number, first 60 AAs:
0.42179
Total prob of N-in:
0.01929
outside
1 - 488
Population Genetic Test Statistics
Pi
205.346523
Theta
129.597137
Tajima's D
3.084825
CLR
0
CSRT
0.980850957452127
Interpretation
Uncertain
