Gene
KWMTBOMO01159
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.466
Sequence
CDS
ATGTCATGGTCCAGACGGCTGGCCGATCCTTCGGCTGGTCGTAGGACCGTCGAGGCGATTCGCCCGGTTCTTGTGAATTGGGTGAATCGTGACAGAGGACGCCTCACTTTCCGGCTCACGCAGGTGCTCACTGGGCATGGTTGCTTCGGTGAGTTCCTGCACCGGATCGGAGCCGAGCCGACGGCAGAGTGCCACCATTGTGGTTGCGGCTTGGACACGGCGGAGCACACGCTCGTCGCCTGCCCCGCATGGGAGGGGTGGCACCGTGTCCTCGTCGCAAAAATAGGAAACGACTTGTCGTTGCCGAGTGTTGTGGCATCGATGCTCGGCGACGACGAGTCGTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGCGTGAGAGACACGCAAGCCCGCCGCCGTCGAGCGGGGGCCAGGGAGGCGGATCTCGCCCAAGCCCTGGCCCTCTAA
Protein
MSWSRRLADPSAGRRTVEAIRPVLVNWVNRDRGRLTFRLTQVLTGHGCFGEFLHRIGAEPTAECHHCGCGLDTAEHTLVACPAWEGWHRVLVAKIGNDLSLPSVVASMLGDDESWKAMLDFCECTISQKEAAGRVRDTQARRRRAGAREADLAQALAL
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
Q5KTM7
Q868Q4
A0A0J7KCI1
A0A0J7KM11
A0A0J7K2M2
A0A2W1BTM2
+ More
A0A3L8D4Y9 A0A0J7MZM4 A0A2W1BSQ5 A0A3L8DDS1 A0A0J7KJB9 A0A2W1C3J5 Q8MY27 A0A2H1VM71 A0A2W1B333 A0A3L8DTK4 A0A3L8D2F4 A0A2W1BXC9 A0A2W1BDU5 Q8MY33 Q8MY31 A0A2H1WBD5 A0A0J7KV16 Q8MY35 A0A0J7N1D9 A0A2W1BY04 A0A0J7KS67 A0A0J7KKS4 A0A0J7KIY3 A0A2H1W776 A0A0J7N7H9 O01419 A0A0J7KCQ3 E2BVR6 A0A0J7L0U0 A0A2H1WNZ5 A0A2H1VYR8 A0A0J7KLT9 A0A0J7KIY5 A0A2H1W4U7 A0A3L8DY91 A0A2H1VTA2 A0A0J7JXN3 A0A0J7KIQ5 A0A0J7MZX3 J9KZ24 A0A0J7N7S8 A0A0J7KJH4 A0A0J7KQK3 A0A3L8D6D2 A0A3L8D4Z4 A0A3L8DIL1 A0A2H1VXT5 A0A2H1WEH8 A0A2H1VVH9 A0A3L8D2G3 A0A0J7KT62 A0A2S2P557 A0A0J7KM81 A0A0J7KJG6 A0A0J7KJ07 A0A3L8DXT9 A0A0J7JZF7 A0A0J7JZ13 A0A2S2P2F0 A0A3L8DDV9 J9KRD0 A0A0J7KP02 A0A0J7KPZ3 A0A0J7KJC5 J9LAC1 A0A1W7R645 A0A2H8TIE5 A0A0J7NCZ4 A0A0J7MW86 A0A0J7KYD2 A0A0J7K3R6 A0A0J7KWU5
A0A3L8D4Y9 A0A0J7MZM4 A0A2W1BSQ5 A0A3L8DDS1 A0A0J7KJB9 A0A2W1C3J5 Q8MY27 A0A2H1VM71 A0A2W1B333 A0A3L8DTK4 A0A3L8D2F4 A0A2W1BXC9 A0A2W1BDU5 Q8MY33 Q8MY31 A0A2H1WBD5 A0A0J7KV16 Q8MY35 A0A0J7N1D9 A0A2W1BY04 A0A0J7KS67 A0A0J7KKS4 A0A0J7KIY3 A0A2H1W776 A0A0J7N7H9 O01419 A0A0J7KCQ3 E2BVR6 A0A0J7L0U0 A0A2H1WNZ5 A0A2H1VYR8 A0A0J7KLT9 A0A0J7KIY5 A0A2H1W4U7 A0A3L8DY91 A0A2H1VTA2 A0A0J7JXN3 A0A0J7KIQ5 A0A0J7MZX3 J9KZ24 A0A0J7N7S8 A0A0J7KJH4 A0A0J7KQK3 A0A3L8D6D2 A0A3L8D4Z4 A0A3L8DIL1 A0A2H1VXT5 A0A2H1WEH8 A0A2H1VVH9 A0A3L8D2G3 A0A0J7KT62 A0A2S2P557 A0A0J7KM81 A0A0J7KJG6 A0A0J7KJ07 A0A3L8DXT9 A0A0J7JZF7 A0A0J7JZ13 A0A2S2P2F0 A0A3L8DDV9 J9KRD0 A0A0J7KP02 A0A0J7KPZ3 A0A0J7KJC5 J9LAC1 A0A1W7R645 A0A2H8TIE5 A0A0J7NCZ4 A0A0J7MW86 A0A0J7KYD2 A0A0J7K3R6 A0A0J7KWU5
EMBL
AB126050
BAD86650.1
AB090825
BAC57926.1
LBMM01009501
KMQ88083.1
+ More
LBMM01005707 KMQ91279.1 LBMM01016102 KMQ84557.1 KZ149922 PZC77701.1 QOIP01000014 RLU15083.1 LBMM01012957 LBMM01012956 KMQ85900.1 KMQ85901.1 KZ149950 PZC76665.1 QOIP01000009 RLU18544.1 LBMM01006538 KMQ90523.1 KZ149896 PZC78733.1 AB078935 BAC06462.1 ODYU01003322 SOQ41921.1 KZ150386 PZC71062.1 QOIP01000004 RLU23632.1 QOIP01000060 RLU14687.1 KZ149918 PZC77857.1 KZ150308 PZC71477.1 AB078930 BAC06454.1 AB078931 BAC06456.1 ODYU01007528 SOQ50378.1 LBMM01003001 KMQ94089.1 AB078929 BAC06452.1 LBMM01012027 KMQ86470.1 PZC77700.1 LBMM01003679 KMQ93242.1 LBMM01006022 KMQ91008.1 LBMM01006909 KMQ90209.1 ODYU01006757 SOQ48893.1 LBMM01008861 KMQ88580.1 D85594 BAA19776.1 LBMM01009494 KMQ88097.1 GL450953 EFN80213.1 LBMM01001564 KMQ96079.1 ODYU01009945 SOQ54696.1 ODYU01005282 SOQ45985.1 LBMM01005733 KMQ91252.1 LBMM01006954 KMQ90166.1 ODYU01006342 SOQ48109.1 QOIP01000002 RLU25407.1 ODYU01004308 SOQ44039.1 LBMM01024024 KMQ82631.1 LBMM01006996 KMQ90129.1 LBMM01012797 KMQ85990.1 ABLF02006132 LBMM01008682 KMQ88705.1 LBMM01006657 KMQ90417.1 LBMM01004272 KMQ92618.1 QOIP01000012 RLU16027.1 QOIP01000013 RLU15211.1 QOIP01000007 RLU20280.1 ODYU01005115 SOQ45661.1 ODYU01008140 SOQ51495.1 ODYU01004693 SOQ44845.1 QOIP01000031 RLU14697.1 LBMM01003452 KMQ93523.1 GGMR01011974 MBY24593.1 LBMM01005634 KMQ91349.1 LBMM01006671 KMQ90407.1 LBMM01006662 KMQ90413.1 QOIP01000003 RLU25053.1 LBMM01019838 KMQ83439.1 LBMM01019839 KMQ83438.1 GGMR01010769 MBY23388.1 QOIP01000010 RLU18098.1 ABLF02026677 ABLF02032994 ABLF02059021 ABLF02064578 LBMM01004906 KMQ92001.1 LBMM01004505 KMQ92346.1 LBMM01006534 KMQ90528.1 ABLF02017307 GEHC01001045 JAV46600.1 GFXV01002090 MBW13895.1 LBMM01006640 KMQ90440.1 LBMM01015657 KMQ84725.1 LBMM01002052 KMQ95336.1 LBMM01015018 KMQ84954.1 LBMM01002480 KMQ94776.1
LBMM01005707 KMQ91279.1 LBMM01016102 KMQ84557.1 KZ149922 PZC77701.1 QOIP01000014 RLU15083.1 LBMM01012957 LBMM01012956 KMQ85900.1 KMQ85901.1 KZ149950 PZC76665.1 QOIP01000009 RLU18544.1 LBMM01006538 KMQ90523.1 KZ149896 PZC78733.1 AB078935 BAC06462.1 ODYU01003322 SOQ41921.1 KZ150386 PZC71062.1 QOIP01000004 RLU23632.1 QOIP01000060 RLU14687.1 KZ149918 PZC77857.1 KZ150308 PZC71477.1 AB078930 BAC06454.1 AB078931 BAC06456.1 ODYU01007528 SOQ50378.1 LBMM01003001 KMQ94089.1 AB078929 BAC06452.1 LBMM01012027 KMQ86470.1 PZC77700.1 LBMM01003679 KMQ93242.1 LBMM01006022 KMQ91008.1 LBMM01006909 KMQ90209.1 ODYU01006757 SOQ48893.1 LBMM01008861 KMQ88580.1 D85594 BAA19776.1 LBMM01009494 KMQ88097.1 GL450953 EFN80213.1 LBMM01001564 KMQ96079.1 ODYU01009945 SOQ54696.1 ODYU01005282 SOQ45985.1 LBMM01005733 KMQ91252.1 LBMM01006954 KMQ90166.1 ODYU01006342 SOQ48109.1 QOIP01000002 RLU25407.1 ODYU01004308 SOQ44039.1 LBMM01024024 KMQ82631.1 LBMM01006996 KMQ90129.1 LBMM01012797 KMQ85990.1 ABLF02006132 LBMM01008682 KMQ88705.1 LBMM01006657 KMQ90417.1 LBMM01004272 KMQ92618.1 QOIP01000012 RLU16027.1 QOIP01000013 RLU15211.1 QOIP01000007 RLU20280.1 ODYU01005115 SOQ45661.1 ODYU01008140 SOQ51495.1 ODYU01004693 SOQ44845.1 QOIP01000031 RLU14697.1 LBMM01003452 KMQ93523.1 GGMR01011974 MBY24593.1 LBMM01005634 KMQ91349.1 LBMM01006671 KMQ90407.1 LBMM01006662 KMQ90413.1 QOIP01000003 RLU25053.1 LBMM01019838 KMQ83439.1 LBMM01019839 KMQ83438.1 GGMR01010769 MBY23388.1 QOIP01000010 RLU18098.1 ABLF02026677 ABLF02032994 ABLF02059021 ABLF02064578 LBMM01004906 KMQ92001.1 LBMM01004505 KMQ92346.1 LBMM01006534 KMQ90528.1 ABLF02017307 GEHC01001045 JAV46600.1 GFXV01002090 MBW13895.1 LBMM01006640 KMQ90440.1 LBMM01015657 KMQ84725.1 LBMM01002052 KMQ95336.1 LBMM01015018 KMQ84954.1 LBMM01002480 KMQ94776.1
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR002123 Plipid/glycerol_acylTrfase
IPR036188 FAD/NAD-bd_sf
IPR012132 GMC_OxRdtase
IPR000172 GMC_OxRdtase_N
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001251 CRAL-TRIO_dom
IPR036865 CRAL-TRIO_dom_sf
IPR036273 CRAL/TRIO_N_dom_sf
IPR006759 Glyco_transf_54
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR002123 Plipid/glycerol_acylTrfase
IPR036188 FAD/NAD-bd_sf
IPR012132 GMC_OxRdtase
IPR000172 GMC_OxRdtase_N
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001251 CRAL-TRIO_dom
IPR036865 CRAL-TRIO_dom_sf
IPR036273 CRAL/TRIO_N_dom_sf
IPR006759 Glyco_transf_54
SUPFAM
Gene 3D
CDD
ProteinModelPortal
Q5KTM7
Q868Q4
A0A0J7KCI1
A0A0J7KM11
A0A0J7K2M2
A0A2W1BTM2
+ More
A0A3L8D4Y9 A0A0J7MZM4 A0A2W1BSQ5 A0A3L8DDS1 A0A0J7KJB9 A0A2W1C3J5 Q8MY27 A0A2H1VM71 A0A2W1B333 A0A3L8DTK4 A0A3L8D2F4 A0A2W1BXC9 A0A2W1BDU5 Q8MY33 Q8MY31 A0A2H1WBD5 A0A0J7KV16 Q8MY35 A0A0J7N1D9 A0A2W1BY04 A0A0J7KS67 A0A0J7KKS4 A0A0J7KIY3 A0A2H1W776 A0A0J7N7H9 O01419 A0A0J7KCQ3 E2BVR6 A0A0J7L0U0 A0A2H1WNZ5 A0A2H1VYR8 A0A0J7KLT9 A0A0J7KIY5 A0A2H1W4U7 A0A3L8DY91 A0A2H1VTA2 A0A0J7JXN3 A0A0J7KIQ5 A0A0J7MZX3 J9KZ24 A0A0J7N7S8 A0A0J7KJH4 A0A0J7KQK3 A0A3L8D6D2 A0A3L8D4Z4 A0A3L8DIL1 A0A2H1VXT5 A0A2H1WEH8 A0A2H1VVH9 A0A3L8D2G3 A0A0J7KT62 A0A2S2P557 A0A0J7KM81 A0A0J7KJG6 A0A0J7KJ07 A0A3L8DXT9 A0A0J7JZF7 A0A0J7JZ13 A0A2S2P2F0 A0A3L8DDV9 J9KRD0 A0A0J7KP02 A0A0J7KPZ3 A0A0J7KJC5 J9LAC1 A0A1W7R645 A0A2H8TIE5 A0A0J7NCZ4 A0A0J7MW86 A0A0J7KYD2 A0A0J7K3R6 A0A0J7KWU5
A0A3L8D4Y9 A0A0J7MZM4 A0A2W1BSQ5 A0A3L8DDS1 A0A0J7KJB9 A0A2W1C3J5 Q8MY27 A0A2H1VM71 A0A2W1B333 A0A3L8DTK4 A0A3L8D2F4 A0A2W1BXC9 A0A2W1BDU5 Q8MY33 Q8MY31 A0A2H1WBD5 A0A0J7KV16 Q8MY35 A0A0J7N1D9 A0A2W1BY04 A0A0J7KS67 A0A0J7KKS4 A0A0J7KIY3 A0A2H1W776 A0A0J7N7H9 O01419 A0A0J7KCQ3 E2BVR6 A0A0J7L0U0 A0A2H1WNZ5 A0A2H1VYR8 A0A0J7KLT9 A0A0J7KIY5 A0A2H1W4U7 A0A3L8DY91 A0A2H1VTA2 A0A0J7JXN3 A0A0J7KIQ5 A0A0J7MZX3 J9KZ24 A0A0J7N7S8 A0A0J7KJH4 A0A0J7KQK3 A0A3L8D6D2 A0A3L8D4Z4 A0A3L8DIL1 A0A2H1VXT5 A0A2H1WEH8 A0A2H1VVH9 A0A3L8D2G3 A0A0J7KT62 A0A2S2P557 A0A0J7KM81 A0A0J7KJG6 A0A0J7KJ07 A0A3L8DXT9 A0A0J7JZF7 A0A0J7JZ13 A0A2S2P2F0 A0A3L8DDV9 J9KRD0 A0A0J7KP02 A0A0J7KPZ3 A0A0J7KJC5 J9LAC1 A0A1W7R645 A0A2H8TIE5 A0A0J7NCZ4 A0A0J7MW86 A0A0J7KYD2 A0A0J7K3R6 A0A0J7KWU5
Ontologies
PANTHER
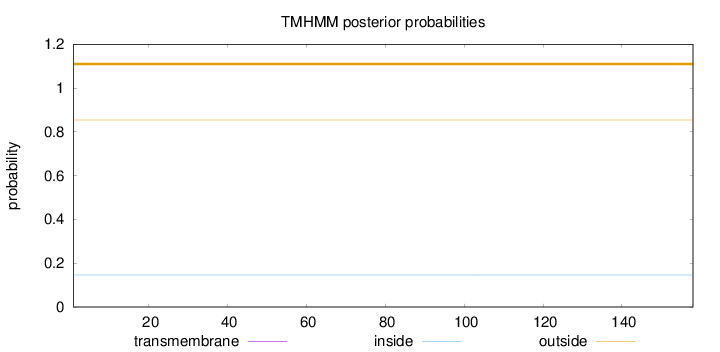
Topology
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00566
Exp number, first 60 AAs:
0.00418
Total prob of N-in:
0.14647
outside
1 - 158
Population Genetic Test Statistics
Pi
32.528918
Theta
24.662235
Tajima's D
0.887537
CLR
8.862084
CSRT
0.633468326583671
Interpretation
Uncertain
