Pre Gene Modal
BGIBMGA007482
Annotation
PREDICTED:_coiled-coil_domain-containing_protein_47_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.916 Nuclear Reliability : 2.446
Sequence
CDS
ATGCGGATAACATTTCTATTGATTGGATTTCTCTGCGGAATACAGTTTGTTTATGGTTTTGAAGACACTACTCTGGAAGATGATGATTTTGCTGAATTTGAACAGTTTGATGCTGAAGATGATACACCCGTCAATGATTATACAGAAGAAGTACCTGATGTGTTGTCACAAAAAGTGAAACCACCACCTAAAGAATTTGAGGCTGTAGTTGAAATTGAAGACGAAATAATATTAGAGGATGAGGACAATGAATTTGAGCATTTCCAAGACCCTGAGGAGTTTGAAGGGTTCCAGGAAACCACTCCTCGTACAATGGAACAACCAAAGATTACTATTTCTAAGGTTCCTATAACGGCTCGACCTCGCTGGGATGCGTACTGGCTGGAGGGCATACTCTGCTGTCTGCTGGCGTCGTACGCGCTCGCCTATGCCATTGGTCGCGCCAAGAACACGTCTATAGCCGTGAACTTTCTGAAGCTACACAAACCGCTTCTGGAGGAGAATTTCACTCTAGTCGGCGAGACTGGCGCGGAGGTGGTGAGCGGCGCGGACGAGCGCGGCTGGAGGCGCGAGGCGGAGCACTGCTTCACGATGTGGTGCAGCGGCCGCAGCTGCTGCGAGGGCATGCTGCTCACGCTCAAACTCATCAAGCGGCAAGACCTCGTCCACGTGCTGCTGGGCGTGGTGCGGCCCAACCCGGACACGCTGCAGATCCGCATAGAGCTCGGCAAAGACGATATGGATCCCTTCGTGTTCTGTATCGCTCAGAAGAAGATAGCCGCGAAACTCGCCAGGGAAATGCACGATTTGAGCGTGTACTGCGGCGAGCGGCGCGCGGGCGAGCGGCTGGGCGTGCCGGGCGCGCTGAGCGTGGTGGCGGAGTGCGCGGAGCCGGGCGCGGCGCTGCTGGACGCGCGCGTGTGCGCCGCGCTGCAGCTGCACCACCGACACCTGCTCTACCTGCACCTCTCCGACAAGTACGCCGGGCCCAAGCACCTCGAGGAGACGGCATCAACGAAGCCCCCGGAGACGGAGCGCGTGATGCTGGTGAGCCTGGCGCTGGGCCCGGACGGCGGCGGGGACGAGCTCCGCCCGCTGCTGCAGCTCGTCTTCTACATCCTCGACAAGCTCAAGAGGTTCAAGCTCAGCAAAGAGGCGTTGGCTAAGTGCGAGAAGCGTCGTCAGAAGGTGGCGGAGGCCTGGCTCCGGGGGGCGCACGCTGCCAGACAAGAGCAGGCCGCTCTAAGGCGGGAGGAAAAACGGAAACAAGAGAAGGAACGCATACTCGCAGAAGACGACCCAGAGAAACAGCGTCGTTGGGAATTAAAAGAGCAAAAGCGCCAGCAGAAACGAAAAACTCCCAAAATGAAACAATTGAAAGTGAAGGCGCTGTGA
Protein
MRITFLLIGFLCGIQFVYGFEDTTLEDDDFAEFEQFDAEDDTPVNDYTEEVPDVLSQKVKPPPKEFEAVVEIEDEIILEDEDNEFEHFQDPEEFEGFQETTPRTMEQPKITISKVPITARPRWDAYWLEGILCCLLASYALAYAIGRAKNTSIAVNFLKLHKPLLEENFTLVGETGAEVVSGADERGWRREAEHCFTMWCSGRSCCEGMLLTLKLIKRQDLVHVLLGVVRPNPDTLQIRIELGKDDMDPFVFCIAQKKIAAKLAREMHDLSVYCGERRAGERLGVPGALSVVAECAEPGAALLDARVCAALQLHHRHLLYLHLSDKYAGPKHLEETASTKPPETERVMLVSLALGPDGGGDELRPLLQLVFYILDKLKRFKLSKEALAKCEKRRQKVAEAWLRGAHAARQEQAALRREEKRKQEKERILAEDDPEKQRRWELKEQKRQQKRKTPKMKQLKVKAL
Summary
Uniprot
H9JD85
A0A2A4JJM2
S4P9X5
A0A1E1WF53
I4DJB7
A0A212F6D0
+ More
A0A0N1IGW9 D6WW36 A0A0T6B515 A0A161MJT5 A0A224XPE1 E0VE15 A0A1S3DCT4 T1IG45 A0A1Y1KK89 A0A2P8YQF1 A0A2J7QZZ3 A0A0A9Z0B6 A0A1B6CV99 A0A1B6LS45 A0A1W4XBB0 A0A1B6M0P8 A0A1B6KQV2 A0A1B6L255 A0A195DHY7 A0A2M3Z507 A0A182TKL0 A0A1Y9GL38 A0A2M3Z4Z9 Q7Q124 W5JC68 A0A182PVQ9 A0A2M4BKZ7 A0A2M4BL94 A0A2M4BL58 A0A232FGA8 A0A195BR26 A0A158P1Q6 A0A182K4X5 A0A195EQM7 A0A2M4ACA6 K7ISZ2 A0A182UU19 A0A182KZ59 A0A182X2F8 F4WKD6 A0A182Y683 A0A182W8P8 A0A182F360 A0A182HC10 A0A182GKD9 A0A151XB08 A0A182NP07 A0A182RWP5 A0A182MVL3 A0A195CLJ8 A0A067REC3 A0A023ET13 Q16GK8 A0A182QPZ5 A0A336M334 B0WPF8 A0A2P6LGH5 U5EJN1 A0A1Q3FCF3 A0A0J7NX27 A0A087UE65 A0A084VSN9 E2BJV1 E2AFH6 T1PC37 A0A182JBI0 A0A023GCT3 E9J3X7 A0A0P4WJH0 A0A0L0BZD2 A0A1I8PWG2 A0A0P4X066 T1FXN9 A0A0C9Q3H5 L7MBF0 A0A0M3QTG4 A0A131YXI7 A0A2A3E5T7 A0A224Y2X6 A0A088AEJ8 N6U4G5 W8CB98 A0A1L8D8W3 A0A0L7QLT3 A0A034VDG4 A0A0K8WIY0 A0A1L8DNQ9
A0A0N1IGW9 D6WW36 A0A0T6B515 A0A161MJT5 A0A224XPE1 E0VE15 A0A1S3DCT4 T1IG45 A0A1Y1KK89 A0A2P8YQF1 A0A2J7QZZ3 A0A0A9Z0B6 A0A1B6CV99 A0A1B6LS45 A0A1W4XBB0 A0A1B6M0P8 A0A1B6KQV2 A0A1B6L255 A0A195DHY7 A0A2M3Z507 A0A182TKL0 A0A1Y9GL38 A0A2M3Z4Z9 Q7Q124 W5JC68 A0A182PVQ9 A0A2M4BKZ7 A0A2M4BL94 A0A2M4BL58 A0A232FGA8 A0A195BR26 A0A158P1Q6 A0A182K4X5 A0A195EQM7 A0A2M4ACA6 K7ISZ2 A0A182UU19 A0A182KZ59 A0A182X2F8 F4WKD6 A0A182Y683 A0A182W8P8 A0A182F360 A0A182HC10 A0A182GKD9 A0A151XB08 A0A182NP07 A0A182RWP5 A0A182MVL3 A0A195CLJ8 A0A067REC3 A0A023ET13 Q16GK8 A0A182QPZ5 A0A336M334 B0WPF8 A0A2P6LGH5 U5EJN1 A0A1Q3FCF3 A0A0J7NX27 A0A087UE65 A0A084VSN9 E2BJV1 E2AFH6 T1PC37 A0A182JBI0 A0A023GCT3 E9J3X7 A0A0P4WJH0 A0A0L0BZD2 A0A1I8PWG2 A0A0P4X066 T1FXN9 A0A0C9Q3H5 L7MBF0 A0A0M3QTG4 A0A131YXI7 A0A2A3E5T7 A0A224Y2X6 A0A088AEJ8 N6U4G5 W8CB98 A0A1L8D8W3 A0A0L7QLT3 A0A034VDG4 A0A0K8WIY0 A0A1L8DNQ9
Pubmed
19121390
23622113
22651552
26354079
22118469
18362917
+ More
19820115 20566863 28004739 29403074 25401762 26823975 12364791 14747013 17210077 20920257 23761445 28648823 21347285 20075255 20966253 21719571 25244985 26483478 24845553 24945155 17510324 24438588 20798317 25315136 21282665 26108605 23254933 25576852 26830274 28797301 23537049 24495485 25348373
19820115 20566863 28004739 29403074 25401762 26823975 12364791 14747013 17210077 20920257 23761445 28648823 21347285 20075255 20966253 21719571 25244985 26483478 24845553 24945155 17510324 24438588 20798317 25315136 21282665 26108605 23254933 25576852 26830274 28797301 23537049 24495485 25348373
EMBL
BABH01036256
BABH01036257
NWSH01001348
PCG71592.1
GAIX01003514
JAA89046.1
+ More
GDQN01011417 GDQN01005455 JAT79637.1 JAT85599.1 AK401385 KQ459249 BAM18007.1 KPJ02352.1 AGBW02010050 OWR49287.1 KQ460366 KPJ15590.1 KQ971361 EFA08193.1 LJIG01009761 KRT82434.1 GEMB01000814 JAS02326.1 GFTR01006535 JAW09891.1 DS235088 EEB11621.1 ACPB03002684 GEZM01083983 JAV60881.1 PYGN01000432 PSN46487.1 NEVH01009067 PNF34147.1 GBHO01023522 GBHO01004917 GBHO01004914 GBHO01004908 GDHC01001480 JAG20082.1 JAG38687.1 JAG38690.1 JAG38696.1 JAQ17149.1 GEDC01019997 JAS17301.1 GEBQ01013502 JAT26475.1 GEBQ01027601 GEBQ01010479 JAT12376.1 JAT29498.1 GEBQ01029248 GEBQ01026167 JAT10729.1 JAT13810.1 GEBQ01022237 JAT17740.1 KQ980824 KYN12456.1 GGFM01002843 MBW23594.1 APCN01001654 GGFM01002835 MBW23586.1 AAAB01008980 EAA13928.4 ADMH02001597 ETN61917.1 GGFJ01004605 MBW53746.1 GGFJ01004592 MBW53733.1 GGFJ01004593 MBW53734.1 NNAY01000247 OXU29732.1 KQ976419 KYM89338.1 ADTU01006753 KQ982021 KYN30518.1 GGFK01005104 MBW38425.1 AAZX01005553 GL888199 EGI65360.1 JXUM01032625 KQ560961 KXJ80198.1 JXUM01069814 KQ562571 KXJ75576.1 KQ982335 KYQ57478.1 AXCM01000351 KQ977595 KYN01601.1 KK852521 KDR22107.1 GAPW01001163 JAC12435.1 CH478268 EAT33366.1 AXCN02001960 UFQS01000338 UFQT01000338 SSX02972.1 SSX23339.1 DS232023 EDS32337.1 MWRG01000098 PRD37681.1 GANO01002176 JAB57695.1 GFDL01009815 JAV25230.1 LBMM01001078 KMQ96970.1 KK119412 KFM75654.1 ATLV01016119 ATLV01016120 KE525057 KFB40983.1 GL448661 EFN84034.1 GL439108 EFN67869.1 KA646331 AFP60960.1 GBBM01003457 JAC31961.1 GL768069 EFZ12454.1 GDRN01040228 GDRN01040227 JAI67166.1 JRES01001205 KNC24604.1 GDRN01040226 JAI67167.1 AMQM01000618 KB096324 ESO06377.1 GBYB01000366 GBYB01000367 GBYB01008648 JAG70133.1 JAG70134.1 JAG78415.1 GACK01004545 JAA60489.1 CP012523 ALC38839.1 GEDV01004638 JAP83919.1 KZ288358 PBC27065.1 GFPF01000762 MAA11908.1 APGK01039945 APGK01039946 KB740975 KB632186 ENN76485.1 ERL89771.1 GAMC01005026 JAC01530.1 GFDF01011175 JAV02909.1 KQ414906 KOC59582.1 GAKP01018810 JAC40142.1 GDHF01001193 JAI51121.1 GFDF01005995 JAV08089.1
GDQN01011417 GDQN01005455 JAT79637.1 JAT85599.1 AK401385 KQ459249 BAM18007.1 KPJ02352.1 AGBW02010050 OWR49287.1 KQ460366 KPJ15590.1 KQ971361 EFA08193.1 LJIG01009761 KRT82434.1 GEMB01000814 JAS02326.1 GFTR01006535 JAW09891.1 DS235088 EEB11621.1 ACPB03002684 GEZM01083983 JAV60881.1 PYGN01000432 PSN46487.1 NEVH01009067 PNF34147.1 GBHO01023522 GBHO01004917 GBHO01004914 GBHO01004908 GDHC01001480 JAG20082.1 JAG38687.1 JAG38690.1 JAG38696.1 JAQ17149.1 GEDC01019997 JAS17301.1 GEBQ01013502 JAT26475.1 GEBQ01027601 GEBQ01010479 JAT12376.1 JAT29498.1 GEBQ01029248 GEBQ01026167 JAT10729.1 JAT13810.1 GEBQ01022237 JAT17740.1 KQ980824 KYN12456.1 GGFM01002843 MBW23594.1 APCN01001654 GGFM01002835 MBW23586.1 AAAB01008980 EAA13928.4 ADMH02001597 ETN61917.1 GGFJ01004605 MBW53746.1 GGFJ01004592 MBW53733.1 GGFJ01004593 MBW53734.1 NNAY01000247 OXU29732.1 KQ976419 KYM89338.1 ADTU01006753 KQ982021 KYN30518.1 GGFK01005104 MBW38425.1 AAZX01005553 GL888199 EGI65360.1 JXUM01032625 KQ560961 KXJ80198.1 JXUM01069814 KQ562571 KXJ75576.1 KQ982335 KYQ57478.1 AXCM01000351 KQ977595 KYN01601.1 KK852521 KDR22107.1 GAPW01001163 JAC12435.1 CH478268 EAT33366.1 AXCN02001960 UFQS01000338 UFQT01000338 SSX02972.1 SSX23339.1 DS232023 EDS32337.1 MWRG01000098 PRD37681.1 GANO01002176 JAB57695.1 GFDL01009815 JAV25230.1 LBMM01001078 KMQ96970.1 KK119412 KFM75654.1 ATLV01016119 ATLV01016120 KE525057 KFB40983.1 GL448661 EFN84034.1 GL439108 EFN67869.1 KA646331 AFP60960.1 GBBM01003457 JAC31961.1 GL768069 EFZ12454.1 GDRN01040228 GDRN01040227 JAI67166.1 JRES01001205 KNC24604.1 GDRN01040226 JAI67167.1 AMQM01000618 KB096324 ESO06377.1 GBYB01000366 GBYB01000367 GBYB01008648 JAG70133.1 JAG70134.1 JAG78415.1 GACK01004545 JAA60489.1 CP012523 ALC38839.1 GEDV01004638 JAP83919.1 KZ288358 PBC27065.1 GFPF01000762 MAA11908.1 APGK01039945 APGK01039946 KB740975 KB632186 ENN76485.1 ERL89771.1 GAMC01005026 JAC01530.1 GFDF01011175 JAV02909.1 KQ414906 KOC59582.1 GAKP01018810 JAC40142.1 GDHF01001193 JAI51121.1 GFDF01005995 JAV08089.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000007266
+ More
UP000009046 UP000079169 UP000015103 UP000245037 UP000235965 UP000192223 UP000078492 UP000075902 UP000075840 UP000007062 UP000000673 UP000075885 UP000215335 UP000078540 UP000005205 UP000075881 UP000078541 UP000002358 UP000075903 UP000075882 UP000076407 UP000007755 UP000076408 UP000075920 UP000069272 UP000069940 UP000249989 UP000075809 UP000075884 UP000075900 UP000075883 UP000078542 UP000027135 UP000008820 UP000075886 UP000002320 UP000036403 UP000054359 UP000030765 UP000008237 UP000000311 UP000095301 UP000075880 UP000037069 UP000095300 UP000015101 UP000092553 UP000242457 UP000005203 UP000019118 UP000030742 UP000053825
UP000009046 UP000079169 UP000015103 UP000245037 UP000235965 UP000192223 UP000078492 UP000075902 UP000075840 UP000007062 UP000000673 UP000075885 UP000215335 UP000078540 UP000005205 UP000075881 UP000078541 UP000002358 UP000075903 UP000075882 UP000076407 UP000007755 UP000076408 UP000075920 UP000069272 UP000069940 UP000249989 UP000075809 UP000075884 UP000075900 UP000075883 UP000078542 UP000027135 UP000008820 UP000075886 UP000002320 UP000036403 UP000054359 UP000030765 UP000008237 UP000000311 UP000095301 UP000075880 UP000037069 UP000095300 UP000015101 UP000092553 UP000242457 UP000005203 UP000019118 UP000030742 UP000053825
PRIDE
SUPFAM
SSF158639
SSF158639
Gene 3D
ProteinModelPortal
H9JD85
A0A2A4JJM2
S4P9X5
A0A1E1WF53
I4DJB7
A0A212F6D0
+ More
A0A0N1IGW9 D6WW36 A0A0T6B515 A0A161MJT5 A0A224XPE1 E0VE15 A0A1S3DCT4 T1IG45 A0A1Y1KK89 A0A2P8YQF1 A0A2J7QZZ3 A0A0A9Z0B6 A0A1B6CV99 A0A1B6LS45 A0A1W4XBB0 A0A1B6M0P8 A0A1B6KQV2 A0A1B6L255 A0A195DHY7 A0A2M3Z507 A0A182TKL0 A0A1Y9GL38 A0A2M3Z4Z9 Q7Q124 W5JC68 A0A182PVQ9 A0A2M4BKZ7 A0A2M4BL94 A0A2M4BL58 A0A232FGA8 A0A195BR26 A0A158P1Q6 A0A182K4X5 A0A195EQM7 A0A2M4ACA6 K7ISZ2 A0A182UU19 A0A182KZ59 A0A182X2F8 F4WKD6 A0A182Y683 A0A182W8P8 A0A182F360 A0A182HC10 A0A182GKD9 A0A151XB08 A0A182NP07 A0A182RWP5 A0A182MVL3 A0A195CLJ8 A0A067REC3 A0A023ET13 Q16GK8 A0A182QPZ5 A0A336M334 B0WPF8 A0A2P6LGH5 U5EJN1 A0A1Q3FCF3 A0A0J7NX27 A0A087UE65 A0A084VSN9 E2BJV1 E2AFH6 T1PC37 A0A182JBI0 A0A023GCT3 E9J3X7 A0A0P4WJH0 A0A0L0BZD2 A0A1I8PWG2 A0A0P4X066 T1FXN9 A0A0C9Q3H5 L7MBF0 A0A0M3QTG4 A0A131YXI7 A0A2A3E5T7 A0A224Y2X6 A0A088AEJ8 N6U4G5 W8CB98 A0A1L8D8W3 A0A0L7QLT3 A0A034VDG4 A0A0K8WIY0 A0A1L8DNQ9
A0A0N1IGW9 D6WW36 A0A0T6B515 A0A161MJT5 A0A224XPE1 E0VE15 A0A1S3DCT4 T1IG45 A0A1Y1KK89 A0A2P8YQF1 A0A2J7QZZ3 A0A0A9Z0B6 A0A1B6CV99 A0A1B6LS45 A0A1W4XBB0 A0A1B6M0P8 A0A1B6KQV2 A0A1B6L255 A0A195DHY7 A0A2M3Z507 A0A182TKL0 A0A1Y9GL38 A0A2M3Z4Z9 Q7Q124 W5JC68 A0A182PVQ9 A0A2M4BKZ7 A0A2M4BL94 A0A2M4BL58 A0A232FGA8 A0A195BR26 A0A158P1Q6 A0A182K4X5 A0A195EQM7 A0A2M4ACA6 K7ISZ2 A0A182UU19 A0A182KZ59 A0A182X2F8 F4WKD6 A0A182Y683 A0A182W8P8 A0A182F360 A0A182HC10 A0A182GKD9 A0A151XB08 A0A182NP07 A0A182RWP5 A0A182MVL3 A0A195CLJ8 A0A067REC3 A0A023ET13 Q16GK8 A0A182QPZ5 A0A336M334 B0WPF8 A0A2P6LGH5 U5EJN1 A0A1Q3FCF3 A0A0J7NX27 A0A087UE65 A0A084VSN9 E2BJV1 E2AFH6 T1PC37 A0A182JBI0 A0A023GCT3 E9J3X7 A0A0P4WJH0 A0A0L0BZD2 A0A1I8PWG2 A0A0P4X066 T1FXN9 A0A0C9Q3H5 L7MBF0 A0A0M3QTG4 A0A131YXI7 A0A2A3E5T7 A0A224Y2X6 A0A088AEJ8 N6U4G5 W8CB98 A0A1L8D8W3 A0A0L7QLT3 A0A034VDG4 A0A0K8WIY0 A0A1L8DNQ9
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.974761

Length:
464
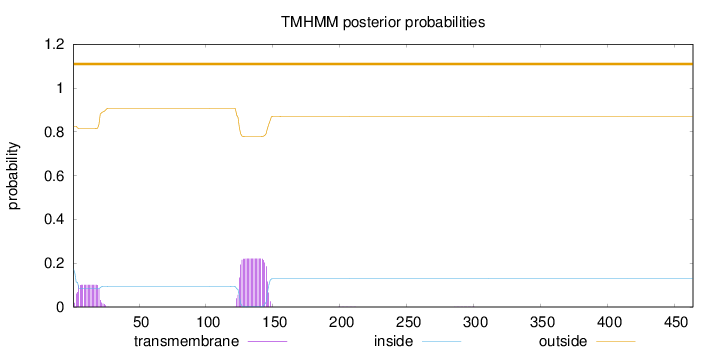
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.61130999999999
Exp number, first 60 AAs:
1.83909
Total prob of N-in:
0.17437
outside
1 - 464
Population Genetic Test Statistics
Pi
221.608763
Theta
140.34433
Tajima's D
1.140066
CLR
0.148797
CSRT
0.701314934253287
Interpretation
Uncertain
