Gene
KWMTBOMO01155 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007458
Annotation
PREDICTED:_lysozyme-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 4.59
Sequence
CDS
ATGGCTTCGGCTGTGATCAGGTTTTGTGCGTTGCTGTTGGTTGCCGGAGTCTGTTTCGCCGATGTTTCGGAACTCCCCCCTGTGACGGAAGTGTGTCTCGGCTGCATTTGCCAGGCTATCTCTGGATGCAAGCAAGGTCTGCAGTGCGAGGGTGAGACGTGCGGCCTCTTCCGCATCACATGGGGCTACTGGGCTGATGCCGGAAAACCCACCATCAATGGTCTCTCACCTGACGCTCCGGATGCCTACTCTAGCTGCACTGTAGACCCGTACTGCGCCGCGCAGACCGTCCAAAACTACATGAAAAGATTTGGCCAGGACTGCAACGGCGACGGAGTGGTCAACTGCTATGACTACATGGCGATCCACAAGAAGGGAGGCTACGGGTGCACCGGCGAACTTCCTTTTAACTATGTAAACGTGTTTAACCAGTGCATCAATGTCTTCGCGCAGTACCGATCTTAA
Protein
MASAVIRFCALLLVAGVCFADVSELPPVTEVCLGCICQAISGCKQGLQCEGETCGLFRITWGYWADAGKPTINGLSPDAPDAYSSCTVDPYCAAQTVQNYMKRFGQDCNGDGVVNCYDYMAIHKKGGYGCTGELPFNYVNVFNQCINVFAQYRS
Summary
Uniprot
H9JD61
A0A346RAE6
A0A1E1WU02
A0A2W1BED9
A0A212F6B8
A0A2A4JE71
+ More
A0A0N1II51 I4DML5 A0A2H1WKR8 I4DIV1 A0A0K8UX72 B4MR88 A0A0L0CMI5 A0A0M4EVW5 A0A0A1XB59 A0A034W495 A0A0C9RMA3 B4LP75 B3NP57 L0AV20 B4HTA5 A0A0J9U3Z4 B4JVL9 Q7JYZ0 A0A0K8VCT2 A0A1A9WK40 B4P5U5 A0A1V1G1I2 Q290K8 B4GBP6 W8C5V6 A0A1W4VCB0 B0XIW2 A0A023EG44 A0A1B6E121 A0A023EJ95 A0A3B0J149 A0A1I8P5L4 B3MD99 A0A2M4ATS5 M9ZTV0 A0A2M4ATK7 A0A1B0BVQ2 A0A1Q3FNH0 A0A182N275 A0A1A9YRF0 A0A2M3ZEU1 A0A1B6C1T8 A0A1B6EIW4 A0A1A9ZP99 W5J4P6 T1PHZ7 A0A182FBZ9 A0A1A9VX79 A0A182WDM6 A0A182R1A9 T1DK01 D3TQQ5 A0A182YAZ0 A0A182LZ45 A0A1B0F0A2 T1DFC2 A0A1B6KZ40 A0A182X5R4 A0A182SN04 A0A182RAL4 A0A336LXM4 A0A182HW82 Q7QHB9 A0A1B6JUP0 A0A182KKY6 A0A1A9VX59 I1XB28 A0A182PJM8 A0A1B6FUZ2 A0A224XMF5 A0A0V0G681 B4LP76 A0A2J7PIA6 B4MR89 A0A1A9XLB9 A0A182UPV2 B4LP77 A0A084WM87 B3DN95 A1ZAL1 A0A1L8E3D0 B4P5U4 B4LP74 A0A1B0BVP8 B3NP56 O96826 B4GBP7 A0A2P8Z5I1 A0A182TFM2 Q290K7 A0A1B0G1Y4 B4QIH1 A0A1A9WK39 Q8SY65
A0A0N1II51 I4DML5 A0A2H1WKR8 I4DIV1 A0A0K8UX72 B4MR88 A0A0L0CMI5 A0A0M4EVW5 A0A0A1XB59 A0A034W495 A0A0C9RMA3 B4LP75 B3NP57 L0AV20 B4HTA5 A0A0J9U3Z4 B4JVL9 Q7JYZ0 A0A0K8VCT2 A0A1A9WK40 B4P5U5 A0A1V1G1I2 Q290K8 B4GBP6 W8C5V6 A0A1W4VCB0 B0XIW2 A0A023EG44 A0A1B6E121 A0A023EJ95 A0A3B0J149 A0A1I8P5L4 B3MD99 A0A2M4ATS5 M9ZTV0 A0A2M4ATK7 A0A1B0BVQ2 A0A1Q3FNH0 A0A182N275 A0A1A9YRF0 A0A2M3ZEU1 A0A1B6C1T8 A0A1B6EIW4 A0A1A9ZP99 W5J4P6 T1PHZ7 A0A182FBZ9 A0A1A9VX79 A0A182WDM6 A0A182R1A9 T1DK01 D3TQQ5 A0A182YAZ0 A0A182LZ45 A0A1B0F0A2 T1DFC2 A0A1B6KZ40 A0A182X5R4 A0A182SN04 A0A182RAL4 A0A336LXM4 A0A182HW82 Q7QHB9 A0A1B6JUP0 A0A182KKY6 A0A1A9VX59 I1XB28 A0A182PJM8 A0A1B6FUZ2 A0A224XMF5 A0A0V0G681 B4LP76 A0A2J7PIA6 B4MR89 A0A1A9XLB9 A0A182UPV2 B4LP77 A0A084WM87 B3DN95 A1ZAL1 A0A1L8E3D0 B4P5U4 B4LP74 A0A1B0BVP8 B3NP56 O96826 B4GBP7 A0A2P8Z5I1 A0A182TFM2 Q290K7 A0A1B0G1Y4 B4QIH1 A0A1A9WK39 Q8SY65
Pubmed
19121390
29910977
28756777
22118469
26354079
22651552
+ More
17994087 26108605 25830018 25348373 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28410430 15632085 24495485 24945155 26483478 23497397 20920257 23761445 25315136 20353571 25244985 24330624 12364791 14747013 17210077 20966253 24438588 29403074
17994087 26108605 25830018 25348373 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28410430 15632085 24495485 24945155 26483478 23497397 20920257 23761445 25315136 20353571 25244985 24330624 12364791 14747013 17210077 20966253 24438588 29403074
EMBL
BABH01036254
BABH01036255
MH200936
AXS59146.1
GDQN01000612
JAT90442.1
+ More
KZ150100 PZC73512.1 AGBW02010050 OWR49285.1 NWSH01001901 PCG69740.1 KQ460366 KPJ15589.1 AK402533 BAM19155.1 ODYU01009331 SOQ53675.1 AK401219 KQ459249 BAM17841.1 KPJ02351.1 GDHF01021118 JAI31196.1 CH963849 EDW74627.1 JRES01000195 KNC33422.1 CP012524 ALC42100.1 GBXI01006066 JAD08226.1 GAKP01010002 JAC48950.1 GBYB01009435 JAG79202.1 CH940648 EDW60184.1 CH954179 EDV55696.1 JX915870 AFZ78836.1 CH480816 EDW48206.1 CM002911 KMY94385.1 CH916375 EDV98487.1 AE013599 AY071664 KX531985 AAF57940.2 AAL49286.1 ANY27795.1 GDHF01015974 JAI36340.1 CM000158 EDW91861.1 FX985424 BAX07437.1 CM000071 EAL25354.2 CH479181 EDW32304.1 GAMC01001482 JAC05074.1 DS233393 EDS29744.1 JXUM01064974 JXUM01064975 JXUM01064976 JXUM01064977 JXUM01064978 JXUM01064979 JXUM01086087 GAPW01005281 KQ563543 KQ562324 JAC08317.1 KXJ73684.1 KXJ76142.1 GEDC01005694 JAS31604.1 GAPW01004567 JAC09031.1 OUUW01000001 SPP74834.1 CH902619 EDV37432.1 GGFK01010791 MBW44112.1 KC355206 AGK40906.1 GGFK01010792 MBW44113.1 JXJN01021433 GFDL01006019 JAV29026.1 GGFM01006247 MBW26998.1 GEDC01029822 JAS07476.1 GECZ01031913 JAS37856.1 ADMH02002203 ETN57835.1 KA647503 AFP62132.1 AXCN02001739 GAMD01001191 JAB00400.1 CCAG010011832 EZ423757 ADD20033.1 AXCM01021388 AJVK01007789 GALA01000677 JAA94175.1 GEBQ01023248 JAT16729.1 UFQT01000174 SSX21107.1 APCN01001474 AY659931 AAAB01008816 AAT51799.1 EAA05103.3 GECU01032937 GECU01004824 JAS74769.1 JAT02883.1 JQ754173 AFI81521.1 GECZ01015785 JAS53984.1 GFTR01002750 JAW13676.1 GECL01002629 JAP03495.1 EDW60185.1 NEVH01025128 PNF16053.1 EDW74628.1 EDW60186.2 ATLV01024393 KE525351 KFB51331.1 BT032883 ACD99447.1 KX671588 AAF57939.1 AOG29613.1 GFDF01001079 JAV13005.1 EDW91860.1 EDW60183.1 JXJN01021428 EDV55695.1 AL031863 CAA21317.1 EDW32305.1 PYGN01000186 PSN51755.1 EAL25356.2 CCAG010007743 CM000362 EDX07418.1 KMY94386.1 AY071760 AAL49382.1
KZ150100 PZC73512.1 AGBW02010050 OWR49285.1 NWSH01001901 PCG69740.1 KQ460366 KPJ15589.1 AK402533 BAM19155.1 ODYU01009331 SOQ53675.1 AK401219 KQ459249 BAM17841.1 KPJ02351.1 GDHF01021118 JAI31196.1 CH963849 EDW74627.1 JRES01000195 KNC33422.1 CP012524 ALC42100.1 GBXI01006066 JAD08226.1 GAKP01010002 JAC48950.1 GBYB01009435 JAG79202.1 CH940648 EDW60184.1 CH954179 EDV55696.1 JX915870 AFZ78836.1 CH480816 EDW48206.1 CM002911 KMY94385.1 CH916375 EDV98487.1 AE013599 AY071664 KX531985 AAF57940.2 AAL49286.1 ANY27795.1 GDHF01015974 JAI36340.1 CM000158 EDW91861.1 FX985424 BAX07437.1 CM000071 EAL25354.2 CH479181 EDW32304.1 GAMC01001482 JAC05074.1 DS233393 EDS29744.1 JXUM01064974 JXUM01064975 JXUM01064976 JXUM01064977 JXUM01064978 JXUM01064979 JXUM01086087 GAPW01005281 KQ563543 KQ562324 JAC08317.1 KXJ73684.1 KXJ76142.1 GEDC01005694 JAS31604.1 GAPW01004567 JAC09031.1 OUUW01000001 SPP74834.1 CH902619 EDV37432.1 GGFK01010791 MBW44112.1 KC355206 AGK40906.1 GGFK01010792 MBW44113.1 JXJN01021433 GFDL01006019 JAV29026.1 GGFM01006247 MBW26998.1 GEDC01029822 JAS07476.1 GECZ01031913 JAS37856.1 ADMH02002203 ETN57835.1 KA647503 AFP62132.1 AXCN02001739 GAMD01001191 JAB00400.1 CCAG010011832 EZ423757 ADD20033.1 AXCM01021388 AJVK01007789 GALA01000677 JAA94175.1 GEBQ01023248 JAT16729.1 UFQT01000174 SSX21107.1 APCN01001474 AY659931 AAAB01008816 AAT51799.1 EAA05103.3 GECU01032937 GECU01004824 JAS74769.1 JAT02883.1 JQ754173 AFI81521.1 GECZ01015785 JAS53984.1 GFTR01002750 JAW13676.1 GECL01002629 JAP03495.1 EDW60185.1 NEVH01025128 PNF16053.1 EDW74628.1 EDW60186.2 ATLV01024393 KE525351 KFB51331.1 BT032883 ACD99447.1 KX671588 AAF57939.1 AOG29613.1 GFDF01001079 JAV13005.1 EDW91860.1 EDW60183.1 JXJN01021428 EDV55695.1 AL031863 CAA21317.1 EDW32305.1 PYGN01000186 PSN51755.1 EAL25356.2 CCAG010007743 CM000362 EDX07418.1 KMY94386.1 AY071760 AAL49382.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000007798
+ More
UP000037069 UP000092553 UP000008792 UP000008711 UP000001292 UP000001070 UP000000803 UP000091820 UP000002282 UP000001819 UP000008744 UP000192221 UP000002320 UP000069940 UP000249989 UP000268350 UP000095300 UP000007801 UP000092460 UP000075884 UP000092443 UP000092445 UP000000673 UP000095301 UP000069272 UP000078200 UP000075920 UP000075886 UP000092444 UP000076408 UP000075883 UP000092462 UP000076407 UP000075901 UP000075900 UP000075840 UP000007062 UP000075882 UP000075885 UP000235965 UP000075903 UP000030765 UP000245037 UP000075902 UP000000304
UP000037069 UP000092553 UP000008792 UP000008711 UP000001292 UP000001070 UP000000803 UP000091820 UP000002282 UP000001819 UP000008744 UP000192221 UP000002320 UP000069940 UP000249989 UP000268350 UP000095300 UP000007801 UP000092460 UP000075884 UP000092443 UP000092445 UP000000673 UP000095301 UP000069272 UP000078200 UP000075920 UP000075886 UP000092444 UP000076408 UP000075883 UP000092462 UP000076407 UP000075901 UP000075900 UP000075840 UP000007062 UP000075882 UP000075885 UP000235965 UP000075903 UP000030765 UP000245037 UP000075902 UP000000304
Interpro
ProteinModelPortal
H9JD61
A0A346RAE6
A0A1E1WU02
A0A2W1BED9
A0A212F6B8
A0A2A4JE71
+ More
A0A0N1II51 I4DML5 A0A2H1WKR8 I4DIV1 A0A0K8UX72 B4MR88 A0A0L0CMI5 A0A0M4EVW5 A0A0A1XB59 A0A034W495 A0A0C9RMA3 B4LP75 B3NP57 L0AV20 B4HTA5 A0A0J9U3Z4 B4JVL9 Q7JYZ0 A0A0K8VCT2 A0A1A9WK40 B4P5U5 A0A1V1G1I2 Q290K8 B4GBP6 W8C5V6 A0A1W4VCB0 B0XIW2 A0A023EG44 A0A1B6E121 A0A023EJ95 A0A3B0J149 A0A1I8P5L4 B3MD99 A0A2M4ATS5 M9ZTV0 A0A2M4ATK7 A0A1B0BVQ2 A0A1Q3FNH0 A0A182N275 A0A1A9YRF0 A0A2M3ZEU1 A0A1B6C1T8 A0A1B6EIW4 A0A1A9ZP99 W5J4P6 T1PHZ7 A0A182FBZ9 A0A1A9VX79 A0A182WDM6 A0A182R1A9 T1DK01 D3TQQ5 A0A182YAZ0 A0A182LZ45 A0A1B0F0A2 T1DFC2 A0A1B6KZ40 A0A182X5R4 A0A182SN04 A0A182RAL4 A0A336LXM4 A0A182HW82 Q7QHB9 A0A1B6JUP0 A0A182KKY6 A0A1A9VX59 I1XB28 A0A182PJM8 A0A1B6FUZ2 A0A224XMF5 A0A0V0G681 B4LP76 A0A2J7PIA6 B4MR89 A0A1A9XLB9 A0A182UPV2 B4LP77 A0A084WM87 B3DN95 A1ZAL1 A0A1L8E3D0 B4P5U4 B4LP74 A0A1B0BVP8 B3NP56 O96826 B4GBP7 A0A2P8Z5I1 A0A182TFM2 Q290K7 A0A1B0G1Y4 B4QIH1 A0A1A9WK39 Q8SY65
A0A0N1II51 I4DML5 A0A2H1WKR8 I4DIV1 A0A0K8UX72 B4MR88 A0A0L0CMI5 A0A0M4EVW5 A0A0A1XB59 A0A034W495 A0A0C9RMA3 B4LP75 B3NP57 L0AV20 B4HTA5 A0A0J9U3Z4 B4JVL9 Q7JYZ0 A0A0K8VCT2 A0A1A9WK40 B4P5U5 A0A1V1G1I2 Q290K8 B4GBP6 W8C5V6 A0A1W4VCB0 B0XIW2 A0A023EG44 A0A1B6E121 A0A023EJ95 A0A3B0J149 A0A1I8P5L4 B3MD99 A0A2M4ATS5 M9ZTV0 A0A2M4ATK7 A0A1B0BVQ2 A0A1Q3FNH0 A0A182N275 A0A1A9YRF0 A0A2M3ZEU1 A0A1B6C1T8 A0A1B6EIW4 A0A1A9ZP99 W5J4P6 T1PHZ7 A0A182FBZ9 A0A1A9VX79 A0A182WDM6 A0A182R1A9 T1DK01 D3TQQ5 A0A182YAZ0 A0A182LZ45 A0A1B0F0A2 T1DFC2 A0A1B6KZ40 A0A182X5R4 A0A182SN04 A0A182RAL4 A0A336LXM4 A0A182HW82 Q7QHB9 A0A1B6JUP0 A0A182KKY6 A0A1A9VX59 I1XB28 A0A182PJM8 A0A1B6FUZ2 A0A224XMF5 A0A0V0G681 B4LP76 A0A2J7PIA6 B4MR89 A0A1A9XLB9 A0A182UPV2 B4LP77 A0A084WM87 B3DN95 A1ZAL1 A0A1L8E3D0 B4P5U4 B4LP74 A0A1B0BVP8 B3NP56 O96826 B4GBP7 A0A2P8Z5I1 A0A182TFM2 Q290K7 A0A1B0G1Y4 B4QIH1 A0A1A9WK39 Q8SY65
PDB
4PJ2
E-value=1.88205e-07,
Score=126
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.999510

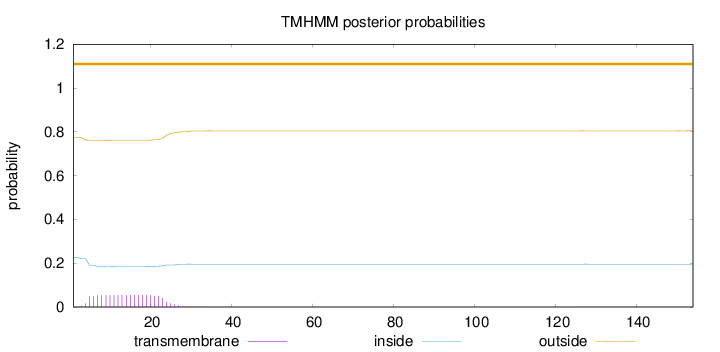
Length:
154
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.11294
Exp number, first 60 AAs:
1.09289
Total prob of N-in:
0.22728
outside
1 - 154
Population Genetic Test Statistics
Pi
207.345748
Theta
176.615851
Tajima's D
0.975284
CLR
0.503156
CSRT
0.653217339133043
Interpretation
Uncertain
