Gene
KWMTBOMO01154
Pre Gene Modal
BGIBMGA007481
Annotation
PREDICTED:_low_density_lipoprotein_receptor_adapter_protein_1-like_[Papilio_polytes]
Full name
Low density lipoprotein receptor adapter protein 1
Alternative Name
Autosomal recessive hypercholesterolemia protein
Location in the cell
Nuclear Reliability : 1.764
Sequence
CDS
ATGGCAACTTTGTTAAGAAAAATGTGGAAAAATCATTCGAAACACAAAAAATTGAGTGAAGAATGGGCATTAGCTGATTGCGAAGGCGAGGGCTGGTGGCGCGAAGTCCGAGATCCGAACAGCAAGATGGACGTCCGCTACGCCGGCTCGGCCCCGGTCGAGCGAGCCGCTTCAGCCCCCGCCACCGTTATTGGTGTACGCACCGCCTTACAGTCTGCCAAAACCCTGAAGAAGAAACCTCAAAAAGTTAACGTCGACATCAACATAAAAGGAATCATTGTGACTGACGCCGAAACAGAGAATAACGTATTAGCGGTTTCCATTTACAGCATCTCGTACTGTTCGGCTGACGTGGCGAATTCTCGAGTTTTCGCTGTTGTGGAGGGAGCGAAGGGAAATGATGAGAACGATTCACATATCGTCCATGTGTTCGTTTGCTCTCAAAGGAAACAAGCACGCGCTCTCGCTCTGTCCTTGGCACACGCTTTCAATGACGCGTATCAGTTATGGCAAGCTAATCAGGCTGGCTCTATGAATTCAAACGCGAAACGGACAGCACCTTGGGTCCGATTCCAGGAAGAATCAGACGAAGAGATTGACGAAGAAGATTGGCAGTCACCGGCTCCGCTTGTCACTTTCGCCTGA
Protein
MATLLRKMWKNHSKHKKLSEEWALADCEGEGWWREVRDPNSKMDVRYAGSAPVERAASAPATVIGVRTALQSAKTLKKKPQKVNVDINIKGIIVTDAETENNVLAVSIYSISYCSADVANSRVFAVVEGAKGNDENDSHIVHVFVCSQRKQARALALSLAHAFNDAYQLWQANQAGSMNSNAKRTAPWVRFQEESDEEIDEEDWQSPAPLVTFA
Summary
Description
Adapter protein (clathrin-associated sorting protein (CLASP)) required for efficient endocytosis of the LDL receptor (LDLR) in polarized cells such as hepatocytes and lymphocytes, but not in non-polarized cells (fibroblasts). May be required for LDL binding and internalization but not for receptor clustering in coated pits. May facilitate the endocytocis of LDLR and LDLR-LDL complexes from coated pits by stabilizing the interaction between the receptor and the structural components of the pits. May also be involved in the internalization of other LDLR family members. Binds to phosphoinositides, which regulate clathrin bud assembly at the cell surface. Required for trafficking of LRP2 to the endocytic recycling compartment which is necessary for LRP2 proteolysis, releasing a tail fragment which translocates to the nucleus and mediates transcriptional repression (By similarity).
Subunit
Interacts (via PID domain) with LDLR (via NPXY motif) (PubMed:12221107). Binds to soluble clathrin trimers (PubMed:12221107). Interacts with AP2B1; the interaction mediates the association with the AP-2 complex (PubMed:12221107). Interacts with VLDLR (By similarity). Interacts with LRP2 (By similarity).
Keywords
3D-structure
Acetylation
Atherosclerosis
Cholesterol metabolism
Complete proteome
Cytoplasm
Disease mutation
Endocytosis
Hyperlipidemia
Lipid metabolism
Phosphoprotein
Polymorphism
Reference proteome
Steroid metabolism
Sterol metabolism
Feature
chain Low density lipoprotein receptor adapter protein 1
sequence variant Probable disease-associated mutation found in patients with hypercholesterolemia; dbSNP:rs752849346.
sequence variant Probable disease-associated mutation found in patients with hypercholesterolemia; dbSNP:rs752849346.
Uniprot
H9JD84
A0A2A4JDH7
I4DJT7
I4DL39
A0A0N1PHF3
A0A2H1X3S5
+ More
A0A194QA01 A0A2W1BPA9 A0A151XDB7 A0A3L8DP13 A0A0P4VQV1 A0A088AW75 A0A146L727 D6WWI6 A0A0C9R9S7 A0A1L8DN98 A0A1B6DA83 A0A0N7Z9W8 A0A2A3EFP4 A0A026VYN1 A0A1B0CNW7 U5ER70 A0A154PEA4 E2AJD2 A0A1Y1N2E7 A0A2M4AC43 A0A2M3ZHQ0 W5JG64 A0A2M3ZHL3 A0A2M4BUB9 A0A2Y9D3L9 A0A0T6BAI5 A0A182THT7 A0A182VM59 A0A2C9GS82 Q7Q4P9 E2B2B4 A0A182F2M3 A0A2M4BUR2 A0A084VBE7 A0A161TJ14 A0A182P4P7 A0A182QJ23 A0A0K8TMI4 A0A182MI92 A0A182ITP6 A0A182RCA1 A0A2M4ABP2 A0A182VTQ2 A0A1Q3G463 A0A182NB42 A0A1B6J8C8 Q16LU8 A0A2P2HY32 A0A1A8D594 T1DR92 A0A1A8BIC0 A0A1A8J4T9 A0A1A7ZBE5 A0A1A8PU27 A0A0S7J3X4 A0A1A8E6S0 A0A1A8VFI2 A0A1A8LKM8 A0A1A8A2V2 A0A1A8PU50 A0A1A8HLC2 A0A1A8JZU0 B3KR97 A0A1A7XSJ6 A0A2J8JNH5 A0A1A8RWB0 A0A1A8ME74 A0A3B1KGN3 F1STU0 A0A1A8RS04 A0A2Y9DLZ6 A0A3L8RVZ5 Q5SW96 G3QXP7 K7CYA0 A0A2J8SIQ6 H2N8I3 H0WK15 A0A3B3DKA6 A0A2K6B5C5 A0A2K5YPA4 A0A2K5LDB3 H9EV37 H9Z7J8 A0A3L7I8I1 A0A2K6G1B2 A0A096NFE1 A0A250YF35 A0A2I2UTI2
A0A194QA01 A0A2W1BPA9 A0A151XDB7 A0A3L8DP13 A0A0P4VQV1 A0A088AW75 A0A146L727 D6WWI6 A0A0C9R9S7 A0A1L8DN98 A0A1B6DA83 A0A0N7Z9W8 A0A2A3EFP4 A0A026VYN1 A0A1B0CNW7 U5ER70 A0A154PEA4 E2AJD2 A0A1Y1N2E7 A0A2M4AC43 A0A2M3ZHQ0 W5JG64 A0A2M3ZHL3 A0A2M4BUB9 A0A2Y9D3L9 A0A0T6BAI5 A0A182THT7 A0A182VM59 A0A2C9GS82 Q7Q4P9 E2B2B4 A0A182F2M3 A0A2M4BUR2 A0A084VBE7 A0A161TJ14 A0A182P4P7 A0A182QJ23 A0A0K8TMI4 A0A182MI92 A0A182ITP6 A0A182RCA1 A0A2M4ABP2 A0A182VTQ2 A0A1Q3G463 A0A182NB42 A0A1B6J8C8 Q16LU8 A0A2P2HY32 A0A1A8D594 T1DR92 A0A1A8BIC0 A0A1A8J4T9 A0A1A7ZBE5 A0A1A8PU27 A0A0S7J3X4 A0A1A8E6S0 A0A1A8VFI2 A0A1A8LKM8 A0A1A8A2V2 A0A1A8PU50 A0A1A8HLC2 A0A1A8JZU0 B3KR97 A0A1A7XSJ6 A0A2J8JNH5 A0A1A8RWB0 A0A1A8ME74 A0A3B1KGN3 F1STU0 A0A1A8RS04 A0A2Y9DLZ6 A0A3L8RVZ5 Q5SW96 G3QXP7 K7CYA0 A0A2J8SIQ6 H2N8I3 H0WK15 A0A3B3DKA6 A0A2K6B5C5 A0A2K5YPA4 A0A2K5LDB3 H9EV37 H9Z7J8 A0A3L7I8I1 A0A2K6G1B2 A0A096NFE1 A0A250YF35 A0A2I2UTI2
Pubmed
19121390
22651552
26354079
28756777
30249741
27129103
+ More
26823975 18362917 19820115 24508170 20798317 28004739 20920257 23761445 12364791 14747013 17210077 24438588 26369729 17510324 14702039 25329095 30282656 11326085 12417523 17974005 16710414 15489334 12221107 12451172 15728179 16903783 18669648 21269460 22814378 23186163 16516836 20124734 22398555 16136131 29451363 25319552 29704459 28087693 17975172
26823975 18362917 19820115 24508170 20798317 28004739 20920257 23761445 12364791 14747013 17210077 24438588 26369729 17510324 14702039 25329095 30282656 11326085 12417523 17974005 16710414 15489334 12221107 12451172 15728179 16903783 18669648 21269460 22814378 23186163 16516836 20124734 22398555 16136131 29451363 25319552 29704459 28087693 17975172
EMBL
BABH01036254
NWSH01001901
PCG69738.1
AK401555
BAM18177.1
AK402007
+ More
BAM18629.1 KQ460366 KPJ15587.1 ODYU01012789 SOQ59274.1 KQ459249 KPJ02348.1 KZ150100 PZC73513.1 KQ982294 KYQ58355.1 QOIP01000006 RLU21972.1 GDKW01003068 JAI53527.1 GDHC01018393 GDHC01017275 GDHC01014395 GDHC01013646 GDHC01011784 JAQ00236.1 JAQ01354.1 JAQ04234.1 JAQ04983.1 JAQ06845.1 KQ971361 EFA08715.1 GBYB01003556 JAG73323.1 GFDF01006158 JAV07926.1 GEDC01014689 JAS22609.1 GDRN01108560 JAI57208.1 KZ288256 PBC30538.1 KK107566 EZA48908.1 AJWK01021047 GANO01003872 JAB55999.1 KQ434874 KZC09754.1 GL439972 EFN66453.1 GEZM01014699 JAV92032.1 GGFK01004857 MBW38178.1 GGFM01007325 MBW28076.1 ADMH02001492 ETN62328.1 GGFM01007306 MBW28057.1 GGFJ01007539 MBW56680.1 LJIG01002608 KRT84332.1 APCN01000743 AAAB01008964 EAA12546.2 GL445130 EFN90151.1 GGFJ01007540 MBW56681.1 ATLV01006583 KE524368 KFB35291.1 GEMB01000243 JAS02872.1 AXCN02000644 GDAI01002230 JAI15373.1 AXCM01000666 GGFK01004885 MBW38206.1 GFDL01000455 JAV34590.1 GECU01012282 JAS95424.1 CH477890 EAT35301.1 IACF01000953 LAB66687.1 HAEA01000008 SBQ28488.1 GAMD01000730 JAB00861.1 HADZ01003406 SBP67347.1 HAED01018187 SBR04632.1 HADY01001647 SBP40132.1 HAEH01008434 SBR84479.1 GBYX01265260 JAO59305.1 HAEA01013690 SBQ42170.1 HADY01016219 HAEJ01017885 SBS58342.1 HAEF01007509 SBR44891.1 HADY01010376 HAEJ01002619 SBP48861.1 HAEG01009537 SBR84768.1 HAEB01000857 HAEC01015228 SBQ83449.1 HAEE01004879 SBR24899.1 AK091219 BAG52309.1 HADW01019682 HADX01012019 SBP21082.1 NBAG03000438 PNI24311.1 HAEI01010214 SBS09982.1 HAEF01013682 SBR54841.1 AEMK02000045 HAEH01018640 SBS08004.1 QUSF01000165 RLV88917.1 AY389348 AL117654 AL606491 BX572623 BC029770 CABD030001780 CABD030001781 AACZ04072242 GABC01006735 GABF01003539 GABD01000691 GABE01006656 JAA04603.1 JAA18606.1 JAA32409.1 JAA38083.1 PNI24310.1 NDHI03003565 PNJ20657.1 ABGA01100205 ABGA01100206 ABGA01203431 ABGA01203432 AAQR03155584 JU322490 AFE66246.1 JU475110 AFH31914.1 RAZU01000087 RLQ74488.1 AHZZ02015522 GFFW01002547 JAV42241.1 AANG04003369
BAM18629.1 KQ460366 KPJ15587.1 ODYU01012789 SOQ59274.1 KQ459249 KPJ02348.1 KZ150100 PZC73513.1 KQ982294 KYQ58355.1 QOIP01000006 RLU21972.1 GDKW01003068 JAI53527.1 GDHC01018393 GDHC01017275 GDHC01014395 GDHC01013646 GDHC01011784 JAQ00236.1 JAQ01354.1 JAQ04234.1 JAQ04983.1 JAQ06845.1 KQ971361 EFA08715.1 GBYB01003556 JAG73323.1 GFDF01006158 JAV07926.1 GEDC01014689 JAS22609.1 GDRN01108560 JAI57208.1 KZ288256 PBC30538.1 KK107566 EZA48908.1 AJWK01021047 GANO01003872 JAB55999.1 KQ434874 KZC09754.1 GL439972 EFN66453.1 GEZM01014699 JAV92032.1 GGFK01004857 MBW38178.1 GGFM01007325 MBW28076.1 ADMH02001492 ETN62328.1 GGFM01007306 MBW28057.1 GGFJ01007539 MBW56680.1 LJIG01002608 KRT84332.1 APCN01000743 AAAB01008964 EAA12546.2 GL445130 EFN90151.1 GGFJ01007540 MBW56681.1 ATLV01006583 KE524368 KFB35291.1 GEMB01000243 JAS02872.1 AXCN02000644 GDAI01002230 JAI15373.1 AXCM01000666 GGFK01004885 MBW38206.1 GFDL01000455 JAV34590.1 GECU01012282 JAS95424.1 CH477890 EAT35301.1 IACF01000953 LAB66687.1 HAEA01000008 SBQ28488.1 GAMD01000730 JAB00861.1 HADZ01003406 SBP67347.1 HAED01018187 SBR04632.1 HADY01001647 SBP40132.1 HAEH01008434 SBR84479.1 GBYX01265260 JAO59305.1 HAEA01013690 SBQ42170.1 HADY01016219 HAEJ01017885 SBS58342.1 HAEF01007509 SBR44891.1 HADY01010376 HAEJ01002619 SBP48861.1 HAEG01009537 SBR84768.1 HAEB01000857 HAEC01015228 SBQ83449.1 HAEE01004879 SBR24899.1 AK091219 BAG52309.1 HADW01019682 HADX01012019 SBP21082.1 NBAG03000438 PNI24311.1 HAEI01010214 SBS09982.1 HAEF01013682 SBR54841.1 AEMK02000045 HAEH01018640 SBS08004.1 QUSF01000165 RLV88917.1 AY389348 AL117654 AL606491 BX572623 BC029770 CABD030001780 CABD030001781 AACZ04072242 GABC01006735 GABF01003539 GABD01000691 GABE01006656 JAA04603.1 JAA18606.1 JAA32409.1 JAA38083.1 PNI24310.1 NDHI03003565 PNJ20657.1 ABGA01100205 ABGA01100206 ABGA01203431 ABGA01203432 AAQR03155584 JU322490 AFE66246.1 JU475110 AFH31914.1 RAZU01000087 RLQ74488.1 AHZZ02015522 GFFW01002547 JAV42241.1 AANG04003369
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000075809
UP000279307
+ More
UP000005203 UP000007266 UP000242457 UP000053097 UP000092461 UP000076502 UP000000311 UP000000673 UP000076407 UP000075902 UP000075903 UP000075840 UP000007062 UP000008237 UP000069272 UP000030765 UP000075885 UP000075886 UP000075883 UP000075880 UP000075900 UP000075920 UP000075884 UP000008820 UP000018467 UP000008227 UP000248480 UP000276834 UP000005640 UP000001519 UP000002277 UP000001595 UP000005225 UP000261560 UP000233120 UP000233140 UP000233060 UP000273346 UP000233160 UP000028761 UP000011712
UP000005203 UP000007266 UP000242457 UP000053097 UP000092461 UP000076502 UP000000311 UP000000673 UP000076407 UP000075902 UP000075903 UP000075840 UP000007062 UP000008237 UP000069272 UP000030765 UP000075885 UP000075886 UP000075883 UP000075880 UP000075900 UP000075920 UP000075884 UP000008820 UP000018467 UP000008227 UP000248480 UP000276834 UP000005640 UP000001519 UP000002277 UP000001595 UP000005225 UP000261560 UP000233120 UP000233140 UP000233060 UP000273346 UP000233160 UP000028761 UP000011712
Interpro
Gene 3D
ProteinModelPortal
H9JD84
A0A2A4JDH7
I4DJT7
I4DL39
A0A0N1PHF3
A0A2H1X3S5
+ More
A0A194QA01 A0A2W1BPA9 A0A151XDB7 A0A3L8DP13 A0A0P4VQV1 A0A088AW75 A0A146L727 D6WWI6 A0A0C9R9S7 A0A1L8DN98 A0A1B6DA83 A0A0N7Z9W8 A0A2A3EFP4 A0A026VYN1 A0A1B0CNW7 U5ER70 A0A154PEA4 E2AJD2 A0A1Y1N2E7 A0A2M4AC43 A0A2M3ZHQ0 W5JG64 A0A2M3ZHL3 A0A2M4BUB9 A0A2Y9D3L9 A0A0T6BAI5 A0A182THT7 A0A182VM59 A0A2C9GS82 Q7Q4P9 E2B2B4 A0A182F2M3 A0A2M4BUR2 A0A084VBE7 A0A161TJ14 A0A182P4P7 A0A182QJ23 A0A0K8TMI4 A0A182MI92 A0A182ITP6 A0A182RCA1 A0A2M4ABP2 A0A182VTQ2 A0A1Q3G463 A0A182NB42 A0A1B6J8C8 Q16LU8 A0A2P2HY32 A0A1A8D594 T1DR92 A0A1A8BIC0 A0A1A8J4T9 A0A1A7ZBE5 A0A1A8PU27 A0A0S7J3X4 A0A1A8E6S0 A0A1A8VFI2 A0A1A8LKM8 A0A1A8A2V2 A0A1A8PU50 A0A1A8HLC2 A0A1A8JZU0 B3KR97 A0A1A7XSJ6 A0A2J8JNH5 A0A1A8RWB0 A0A1A8ME74 A0A3B1KGN3 F1STU0 A0A1A8RS04 A0A2Y9DLZ6 A0A3L8RVZ5 Q5SW96 G3QXP7 K7CYA0 A0A2J8SIQ6 H2N8I3 H0WK15 A0A3B3DKA6 A0A2K6B5C5 A0A2K5YPA4 A0A2K5LDB3 H9EV37 H9Z7J8 A0A3L7I8I1 A0A2K6G1B2 A0A096NFE1 A0A250YF35 A0A2I2UTI2
A0A194QA01 A0A2W1BPA9 A0A151XDB7 A0A3L8DP13 A0A0P4VQV1 A0A088AW75 A0A146L727 D6WWI6 A0A0C9R9S7 A0A1L8DN98 A0A1B6DA83 A0A0N7Z9W8 A0A2A3EFP4 A0A026VYN1 A0A1B0CNW7 U5ER70 A0A154PEA4 E2AJD2 A0A1Y1N2E7 A0A2M4AC43 A0A2M3ZHQ0 W5JG64 A0A2M3ZHL3 A0A2M4BUB9 A0A2Y9D3L9 A0A0T6BAI5 A0A182THT7 A0A182VM59 A0A2C9GS82 Q7Q4P9 E2B2B4 A0A182F2M3 A0A2M4BUR2 A0A084VBE7 A0A161TJ14 A0A182P4P7 A0A182QJ23 A0A0K8TMI4 A0A182MI92 A0A182ITP6 A0A182RCA1 A0A2M4ABP2 A0A182VTQ2 A0A1Q3G463 A0A182NB42 A0A1B6J8C8 Q16LU8 A0A2P2HY32 A0A1A8D594 T1DR92 A0A1A8BIC0 A0A1A8J4T9 A0A1A7ZBE5 A0A1A8PU27 A0A0S7J3X4 A0A1A8E6S0 A0A1A8VFI2 A0A1A8LKM8 A0A1A8A2V2 A0A1A8PU50 A0A1A8HLC2 A0A1A8JZU0 B3KR97 A0A1A7XSJ6 A0A2J8JNH5 A0A1A8RWB0 A0A1A8ME74 A0A3B1KGN3 F1STU0 A0A1A8RS04 A0A2Y9DLZ6 A0A3L8RVZ5 Q5SW96 G3QXP7 K7CYA0 A0A2J8SIQ6 H2N8I3 H0WK15 A0A3B3DKA6 A0A2K6B5C5 A0A2K5YPA4 A0A2K5LDB3 H9EV37 H9Z7J8 A0A3L7I8I1 A0A2K6G1B2 A0A096NFE1 A0A250YF35 A0A2I2UTI2
PDB
3SO6
E-value=2.07485e-18,
Score=223
Ontologies
GO
GO:0070776
GO:0016573
GO:0008285
GO:0006355
GO:0006289
GO:0000439
GO:1904707
GO:0001540
GO:0042982
GO:0001784
GO:0009898
GO:0042632
GO:1905602
GO:0035612
GO:0034383
GO:0035615
GO:0031623
GO:0030159
GO:0009925
GO:0035650
GO:0005769
GO:0005883
GO:0050750
GO:0005546
GO:1905581
GO:0043393
GO:0090118
GO:0071345
GO:0055037
GO:0005829
GO:1903076
GO:0030301
GO:0090205
GO:0030424
GO:0035591
GO:0030276
GO:0005886
GO:0061024
GO:0048260
GO:0006898
GO:0030665
GO:0008203
GO:0005515
GO:0008270
GO:0016831
GO:0004163
GO:0019287
GO:0005634
GO:0006334
GO:0003779
GO:0016021
PANTHER
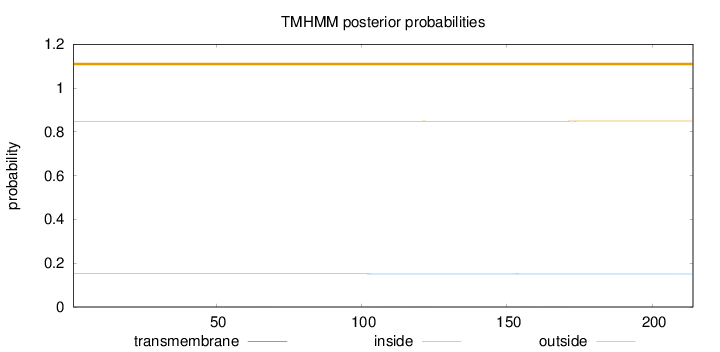
Topology
Subcellular location
Cytoplasm
Length:
214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01797
Exp number, first 60 AAs:
0.00264
Total prob of N-in:
0.15151
outside
1 - 214
Population Genetic Test Statistics
Pi
214.402955
Theta
170.302942
Tajima's D
0.673728
CLR
0.023915
CSRT
0.564421778911054
Interpretation
Uncertain
