Gene
KWMTBOMO01153
Pre Gene Modal
BGIBMGA007480
Annotation
hypothetical_protein_KGM_06329_[Danaus_plexippus]
Full name
Inhibitor of growth protein
Location in the cell
Nuclear Reliability : 3.973
Sequence
CDS
ATGACTTCTGCTCTTTATTTGGAGCATTATTTGGACAGTCTACAGCACTTGCCGATTGAACTGCAAAGAAACTTCAAGTTGATGAGAGATTTGGATGACCGCGCTCACGGGTTAATGAGAACCATTGACCTTATGGCCGATGAGTTGCTTCCAAGTATTCCAAAAATGGATGAAGAAACAAAAAAAGAAAAGGTGAACACTATTCAAGGATTATTCAATAAAGCTAAAGAGTATGGAGACGATAAAGTGCAACTTGCAATTCAGACCTACGAATTAGTAGACAAGCATATACGTAGACTTGATTCTGACCTCGCGCGTTTTGAGTCTGAAATACAAGAGAAAGTAATGAGTGCTCGTGCAGCTCAACAGGCTGCTGCAGATCAGGAAACAAATCCGGCCACAGCTAAAAAAGGTAGGAAAAAGCATAAAGGTACTGAGAAAGCAGCTGCTACGACTGGTAAAAAGAAGCGTGCCGGGGCCTCCAGTGAAGAAGACGCTGTCGCATCCGGAAGGTCAGCTGCGAAGAAGAAAGTTCAGCGTAAAGGTACCACCACCACAACAAATGTTACAAAAGAACAGGAAGAGACTGCCGATTTGGATTCAGTGGCCGGCATGGCACACCCCAGTGATGTGCTAGACATGCCGGTGGATCCTAATGAACCAACATACTGTCTGTGTCATCAAGTTTCATATGGTGAAATGATTGGTTGTGATAACCCAGATTGCCCAATTGAGTGGTTCCATTTTGCTTGTGTTGATCTGAAAATTAAACCTAAGGGTAAATGGTACTGCCCTAAGTGCACTCAGGATAGAAAAAAGAAATGA
Protein
MTSALYLEHYLDSLQHLPIELQRNFKLMRDLDDRAHGLMRTIDLMADELLPSIPKMDEETKKEKVNTIQGLFNKAKEYGDDKVQLAIQTYELVDKHIRRLDSDLARFESEIQEKVMSARAAQQAAADQETNPATAKKGRKKHKGTEKAAATTGKKKRAGASSEEDAVASGRSAAKKKVQRKGTTTTTNVTKEQEETADLDSVAGMAHPSDVLDMPVDPNEPTYCLCHQVSYGEMIGCDNPDCPIEWFHFACVDLKIKPKGKWYCPKCTQDRKKK
Summary
Subunit
Interacts with H3K4me3 and to a lesser extent with H3K4me2.
Similarity
Belongs to the ING family.
Feature
chain Inhibitor of growth protein
Uniprot
H9JD83
A0A212F0H1
A0A2H1X030
A0A2A4JCB6
A0A2W1BIJ6
A0A1E1WHN5
+ More
A0A0L7L1T8 S4NVC9 A0A1W4XH58 A0A139WIF6 D6WK90 A0A1Y1MD45 E2ATY9 A0A2J7QYE2 A0A0J7KG56 J3JTC5 A0A1B6MED8 A0A0N0BFN7 A0A1B6JXE8 A0A182K952 A0A3L8DM44 A0A182HR91 A0A182TJU2 A0A182XGY4 Q7Q136 A0A182KZ45 A0A182VMW5 A0A1B6FH13 A0A151I681 A0A158P1A6 A0A182R159 E0VRC5 A0A182W8R2 A0A0L7RAQ4 A0A182M841 A0A087ZP23 A0A0P4W506 R4G4N9 A0A182P7Z5 A0A084VSR5 A0A2M4CPK5 A0A1B6DWI8 A0A182F555 A0A2M4BWN7 A0A069DQJ6 A0A2M3ZIK8 R4WDF9 A0A2M3ZAN0 A0A2M4AVK6 A0A1L8DX37 A0A0V0GCL2 A0A182Y6A0 A0A224XVW6 A0A0K8TL82 A0A2M4CPD2 W5J7N8 E9IKJ3 B4M967 A0A0Q9VYN3 B3MV97 B4JP81 A0A0K8WAC5 A0A1I8PEN5 A0A1I8PEV4 A0A034WJK6 B4KK64 A0A1I8PEY4 A0A0Q9X4M1 Q29P94 B5DV76 A0A1W4V8G4 A0A182IVQ1 A0A067R4A6 A0A3B0JXJ1 A0A151K0F8 A0A195DL09 A0A151XHC2 A0A0M4ESD8 A0A151IGI7 T1P9R5 A0A1I8PEX2 A0A336MJI8 A0A1I8PEX7 A0A1I8PEZ0 A0A336LG69 A0A1I8PEP2 F4WA34 A0A0R3P1Y9 A0A0R3NQW2 A0A0N1IPG8 A0A194QA01 A0A0C9R1U6 A0A0K8TZ83 A0A0P5LX46 W8CBK3 A0A0P6BZR1 A0A0P5FNR3 A0A0P8ZHY9 A0A1W4UV59
A0A0L7L1T8 S4NVC9 A0A1W4XH58 A0A139WIF6 D6WK90 A0A1Y1MD45 E2ATY9 A0A2J7QYE2 A0A0J7KG56 J3JTC5 A0A1B6MED8 A0A0N0BFN7 A0A1B6JXE8 A0A182K952 A0A3L8DM44 A0A182HR91 A0A182TJU2 A0A182XGY4 Q7Q136 A0A182KZ45 A0A182VMW5 A0A1B6FH13 A0A151I681 A0A158P1A6 A0A182R159 E0VRC5 A0A182W8R2 A0A0L7RAQ4 A0A182M841 A0A087ZP23 A0A0P4W506 R4G4N9 A0A182P7Z5 A0A084VSR5 A0A2M4CPK5 A0A1B6DWI8 A0A182F555 A0A2M4BWN7 A0A069DQJ6 A0A2M3ZIK8 R4WDF9 A0A2M3ZAN0 A0A2M4AVK6 A0A1L8DX37 A0A0V0GCL2 A0A182Y6A0 A0A224XVW6 A0A0K8TL82 A0A2M4CPD2 W5J7N8 E9IKJ3 B4M967 A0A0Q9VYN3 B3MV97 B4JP81 A0A0K8WAC5 A0A1I8PEN5 A0A1I8PEV4 A0A034WJK6 B4KK64 A0A1I8PEY4 A0A0Q9X4M1 Q29P94 B5DV76 A0A1W4V8G4 A0A182IVQ1 A0A067R4A6 A0A3B0JXJ1 A0A151K0F8 A0A195DL09 A0A151XHC2 A0A0M4ESD8 A0A151IGI7 T1P9R5 A0A1I8PEX2 A0A336MJI8 A0A1I8PEX7 A0A1I8PEZ0 A0A336LG69 A0A1I8PEP2 F4WA34 A0A0R3P1Y9 A0A0R3NQW2 A0A0N1IPG8 A0A194QA01 A0A0C9R1U6 A0A0K8TZ83 A0A0P5LX46 W8CBK3 A0A0P6BZR1 A0A0P5FNR3 A0A0P8ZHY9 A0A1W4UV59
Pubmed
19121390
22118469
28756777
26227816
23622113
18362917
+ More
19820115 28004739 20798317 22516182 23537049 30249741 12364791 14747013 17210077 20966253 21347285 20566863 27129103 24438588 26334808 23691247 25244985 26369729 20920257 23761445 21282665 17994087 25348373 15632085 24845553 25315136 21719571 26354079 24495485
19820115 28004739 20798317 22516182 23537049 30249741 12364791 14747013 17210077 20966253 21347285 20566863 27129103 24438588 26334808 23691247 25244985 26369729 20920257 23761445 21282665 17994087 25348373 15632085 24845553 25315136 21719571 26354079 24495485
EMBL
BABH01036254
AGBW02011090
OWR47230.1
ODYU01012382
SOQ58663.1
NWSH01001901
+ More
PCG69737.1 KZ150100 PZC73514.1 GDQN01004549 JAT86505.1 JTDY01003604 KOB69276.1 GAIX01011576 JAA80984.1 KQ971342 KYB27625.1 EFA03615.2 GEZM01034556 GEZM01034554 JAV83732.1 GL442747 EFN63120.1 NEVH01009093 PNF33610.1 LBMM01008059 KMQ89181.1 APGK01034109 BT126481 KB740904 KB632006 AEE61445.1 ENN78442.1 ERL87953.1 GEBQ01005686 JAT34291.1 KQ435798 KOX73348.1 GECU01003819 JAT03888.1 QOIP01000006 RLU21396.1 APCN01001659 AAAB01008980 EAA14095.3 GECZ01020267 JAS49502.1 KQ976402 KYM92286.1 ADTU01006170 ADTU01006171 AXCN02001964 DS235465 EEB15931.1 KQ414618 KOC67944.1 AXCM01000709 GDKW01000134 JAI56461.1 ACPB03000298 GAHY01000611 JAA76899.1 ATLV01016137 KE525057 KFB41009.1 GGFL01003017 MBW67195.1 GEDC01007244 JAS30054.1 GGFJ01008283 MBW57424.1 GBGD01002551 JAC86338.1 GGFM01007588 MBW28339.1 AK417648 BAN20863.1 GGFM01004779 MBW25530.1 GGFK01011421 MBW44742.1 GFDF01003110 JAV10974.1 GECL01000483 JAP05641.1 GFTR01004277 JAW12149.1 GDAI01002682 JAI14921.1 GGFL01003018 MBW67196.1 ADMH02002089 ETN59393.1 GL763991 EFZ18893.1 CH940654 EDW57743.1 KRF77930.1 CH902624 EDV33162.1 CH916372 EDV98711.1 GDHF01004272 JAI48042.1 GAKP01003221 JAC55731.1 CH933807 EDW11582.1 KRG02781.1 CH379058 EAL34399.2 CH675784 EDY71817.1 KK852714 KDR17905.1 OUUW01000010 SPP85783.1 KQ981301 KYN43055.1 KQ980762 KYN13570.1 KQ982138 KYQ59688.1 CP012523 ALC39970.1 KQ977743 KYN00173.1 KA644688 AFP59317.1 UFQT01001120 SSX29003.1 UFQS01002029 UFQT01002029 SSX13002.1 SSX32442.1 GL888037 EGI68964.1 KRT05743.1 KRT03463.1 KQ460366 KPJ15586.1 KQ459249 KPJ02348.1 GBYB01006792 JAG76559.1 GDHF01032540 JAI19774.1 GDIQ01166836 LRGB01000966 JAK84889.1 KZS14329.1 GAMC01004716 JAC01840.1 GDIP01011789 JAM91926.1 GDIQ01255109 JAJ96615.1 KPU74321.1
PCG69737.1 KZ150100 PZC73514.1 GDQN01004549 JAT86505.1 JTDY01003604 KOB69276.1 GAIX01011576 JAA80984.1 KQ971342 KYB27625.1 EFA03615.2 GEZM01034556 GEZM01034554 JAV83732.1 GL442747 EFN63120.1 NEVH01009093 PNF33610.1 LBMM01008059 KMQ89181.1 APGK01034109 BT126481 KB740904 KB632006 AEE61445.1 ENN78442.1 ERL87953.1 GEBQ01005686 JAT34291.1 KQ435798 KOX73348.1 GECU01003819 JAT03888.1 QOIP01000006 RLU21396.1 APCN01001659 AAAB01008980 EAA14095.3 GECZ01020267 JAS49502.1 KQ976402 KYM92286.1 ADTU01006170 ADTU01006171 AXCN02001964 DS235465 EEB15931.1 KQ414618 KOC67944.1 AXCM01000709 GDKW01000134 JAI56461.1 ACPB03000298 GAHY01000611 JAA76899.1 ATLV01016137 KE525057 KFB41009.1 GGFL01003017 MBW67195.1 GEDC01007244 JAS30054.1 GGFJ01008283 MBW57424.1 GBGD01002551 JAC86338.1 GGFM01007588 MBW28339.1 AK417648 BAN20863.1 GGFM01004779 MBW25530.1 GGFK01011421 MBW44742.1 GFDF01003110 JAV10974.1 GECL01000483 JAP05641.1 GFTR01004277 JAW12149.1 GDAI01002682 JAI14921.1 GGFL01003018 MBW67196.1 ADMH02002089 ETN59393.1 GL763991 EFZ18893.1 CH940654 EDW57743.1 KRF77930.1 CH902624 EDV33162.1 CH916372 EDV98711.1 GDHF01004272 JAI48042.1 GAKP01003221 JAC55731.1 CH933807 EDW11582.1 KRG02781.1 CH379058 EAL34399.2 CH675784 EDY71817.1 KK852714 KDR17905.1 OUUW01000010 SPP85783.1 KQ981301 KYN43055.1 KQ980762 KYN13570.1 KQ982138 KYQ59688.1 CP012523 ALC39970.1 KQ977743 KYN00173.1 KA644688 AFP59317.1 UFQT01001120 SSX29003.1 UFQS01002029 UFQT01002029 SSX13002.1 SSX32442.1 GL888037 EGI68964.1 KRT05743.1 KRT03463.1 KQ460366 KPJ15586.1 KQ459249 KPJ02348.1 GBYB01006792 JAG76559.1 GDHF01032540 JAI19774.1 GDIQ01166836 LRGB01000966 JAK84889.1 KZS14329.1 GAMC01004716 JAC01840.1 GDIP01011789 JAM91926.1 GDIQ01255109 JAJ96615.1 KPU74321.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000037510
UP000192223
UP000007266
+ More
UP000000311 UP000235965 UP000036403 UP000019118 UP000030742 UP000053105 UP000075881 UP000279307 UP000075840 UP000075902 UP000076407 UP000007062 UP000075882 UP000075903 UP000078540 UP000005205 UP000075886 UP000009046 UP000075920 UP000053825 UP000075883 UP000005203 UP000015103 UP000075885 UP000030765 UP000069272 UP000076408 UP000000673 UP000008792 UP000007801 UP000001070 UP000095300 UP000009192 UP000001819 UP000192221 UP000075880 UP000027135 UP000268350 UP000078541 UP000078492 UP000075809 UP000092553 UP000078542 UP000095301 UP000007755 UP000053240 UP000053268 UP000076858
UP000000311 UP000235965 UP000036403 UP000019118 UP000030742 UP000053105 UP000075881 UP000279307 UP000075840 UP000075902 UP000076407 UP000007062 UP000075882 UP000075903 UP000078540 UP000005205 UP000075886 UP000009046 UP000075920 UP000053825 UP000075883 UP000005203 UP000015103 UP000075885 UP000030765 UP000069272 UP000076408 UP000000673 UP000008792 UP000007801 UP000001070 UP000095300 UP000009192 UP000001819 UP000192221 UP000075880 UP000027135 UP000268350 UP000078541 UP000078492 UP000075809 UP000092553 UP000078542 UP000095301 UP000007755 UP000053240 UP000053268 UP000076858
PRIDE
Interpro
IPR028651
ING_fam
+ More
IPR028647 ING4
IPR019786 Zinc_finger_PHD-type_CS
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR024610 ING_N_histone_binding
IPR011011 Znf_FYVE_PHD
IPR028638 ING5
IPR011009 Kinase-like_dom_sf
IPR000687 RIO_kinase
IPR018935 RIO_kinase_CS
IPR006020 PTB/PI_dom
IPR011993 PH-like_dom_sf
IPR028647 ING4
IPR019786 Zinc_finger_PHD-type_CS
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR024610 ING_N_histone_binding
IPR011011 Znf_FYVE_PHD
IPR028638 ING5
IPR011009 Kinase-like_dom_sf
IPR000687 RIO_kinase
IPR018935 RIO_kinase_CS
IPR006020 PTB/PI_dom
IPR011993 PH-like_dom_sf
Gene 3D
ProteinModelPortal
H9JD83
A0A212F0H1
A0A2H1X030
A0A2A4JCB6
A0A2W1BIJ6
A0A1E1WHN5
+ More
A0A0L7L1T8 S4NVC9 A0A1W4XH58 A0A139WIF6 D6WK90 A0A1Y1MD45 E2ATY9 A0A2J7QYE2 A0A0J7KG56 J3JTC5 A0A1B6MED8 A0A0N0BFN7 A0A1B6JXE8 A0A182K952 A0A3L8DM44 A0A182HR91 A0A182TJU2 A0A182XGY4 Q7Q136 A0A182KZ45 A0A182VMW5 A0A1B6FH13 A0A151I681 A0A158P1A6 A0A182R159 E0VRC5 A0A182W8R2 A0A0L7RAQ4 A0A182M841 A0A087ZP23 A0A0P4W506 R4G4N9 A0A182P7Z5 A0A084VSR5 A0A2M4CPK5 A0A1B6DWI8 A0A182F555 A0A2M4BWN7 A0A069DQJ6 A0A2M3ZIK8 R4WDF9 A0A2M3ZAN0 A0A2M4AVK6 A0A1L8DX37 A0A0V0GCL2 A0A182Y6A0 A0A224XVW6 A0A0K8TL82 A0A2M4CPD2 W5J7N8 E9IKJ3 B4M967 A0A0Q9VYN3 B3MV97 B4JP81 A0A0K8WAC5 A0A1I8PEN5 A0A1I8PEV4 A0A034WJK6 B4KK64 A0A1I8PEY4 A0A0Q9X4M1 Q29P94 B5DV76 A0A1W4V8G4 A0A182IVQ1 A0A067R4A6 A0A3B0JXJ1 A0A151K0F8 A0A195DL09 A0A151XHC2 A0A0M4ESD8 A0A151IGI7 T1P9R5 A0A1I8PEX2 A0A336MJI8 A0A1I8PEX7 A0A1I8PEZ0 A0A336LG69 A0A1I8PEP2 F4WA34 A0A0R3P1Y9 A0A0R3NQW2 A0A0N1IPG8 A0A194QA01 A0A0C9R1U6 A0A0K8TZ83 A0A0P5LX46 W8CBK3 A0A0P6BZR1 A0A0P5FNR3 A0A0P8ZHY9 A0A1W4UV59
A0A0L7L1T8 S4NVC9 A0A1W4XH58 A0A139WIF6 D6WK90 A0A1Y1MD45 E2ATY9 A0A2J7QYE2 A0A0J7KG56 J3JTC5 A0A1B6MED8 A0A0N0BFN7 A0A1B6JXE8 A0A182K952 A0A3L8DM44 A0A182HR91 A0A182TJU2 A0A182XGY4 Q7Q136 A0A182KZ45 A0A182VMW5 A0A1B6FH13 A0A151I681 A0A158P1A6 A0A182R159 E0VRC5 A0A182W8R2 A0A0L7RAQ4 A0A182M841 A0A087ZP23 A0A0P4W506 R4G4N9 A0A182P7Z5 A0A084VSR5 A0A2M4CPK5 A0A1B6DWI8 A0A182F555 A0A2M4BWN7 A0A069DQJ6 A0A2M3ZIK8 R4WDF9 A0A2M3ZAN0 A0A2M4AVK6 A0A1L8DX37 A0A0V0GCL2 A0A182Y6A0 A0A224XVW6 A0A0K8TL82 A0A2M4CPD2 W5J7N8 E9IKJ3 B4M967 A0A0Q9VYN3 B3MV97 B4JP81 A0A0K8WAC5 A0A1I8PEN5 A0A1I8PEV4 A0A034WJK6 B4KK64 A0A1I8PEY4 A0A0Q9X4M1 Q29P94 B5DV76 A0A1W4V8G4 A0A182IVQ1 A0A067R4A6 A0A3B0JXJ1 A0A151K0F8 A0A195DL09 A0A151XHC2 A0A0M4ESD8 A0A151IGI7 T1P9R5 A0A1I8PEX2 A0A336MJI8 A0A1I8PEX7 A0A1I8PEZ0 A0A336LG69 A0A1I8PEP2 F4WA34 A0A0R3P1Y9 A0A0R3NQW2 A0A0N1IPG8 A0A194QA01 A0A0C9R1U6 A0A0K8TZ83 A0A0P5LX46 W8CBK3 A0A0P6BZR1 A0A0P5FNR3 A0A0P8ZHY9 A0A1W4UV59
PDB
1WEU
E-value=1.37914e-33,
Score=356
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
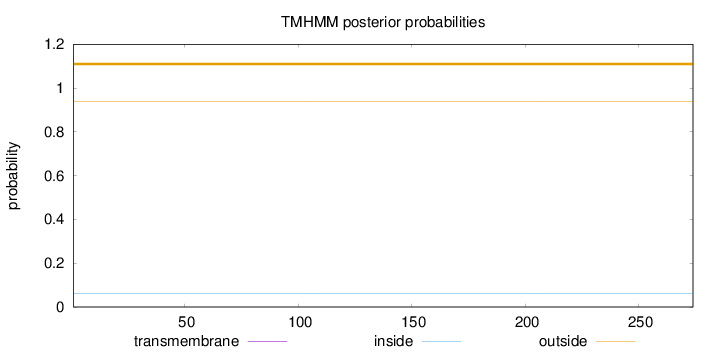
Length:
274
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06031
outside
1 - 274
Population Genetic Test Statistics
Pi
254.607203
Theta
224.483929
Tajima's D
0.492966
CLR
98.060283
CSRT
0.513324333783311
Interpretation
Uncertain
