Pre Gene Modal
BGIBMGA007459
Annotation
diphosphomevalonate_decarboxylase_[Bombyx_mori]
Full name
Diphosphomevalonate decarboxylase
Alternative Name
Mevalonate (diphospho)decarboxylase
Mevalonate pyrophosphate decarboxylase
Mevalonate pyrophosphate decarboxylase
Location in the cell
Cytoplasmic Reliability : 0.965
Sequence
CDS
ATGAGCAACATAGTTACAGTTATTGCTCCAGTAAATATTGCAGTAATAAAATACTGGGGAAAACGTGACGAGAAATTAATTTTACCGTTAAATGATTCCGTTAGTGCTACTTTCGATACAAGTGTTATGTGTGCTAAAACATCGGTATCTACTCATCCGGATTTCGTAGAAGATCAGATTTGGTTAAATGGAAAAGAAGAGTCATTTTCGAATCCAAGATTACAAAACTGCCTCAGGGAGATAAAATCACGAGCTGTTGCAGAGAAAACGATTGCTGAAGATGTACTATCATGGAAAGTTCATGTTTCTTCAGAGAACAATTTTCCAACCGCTGCTGGTTTAGCATCCTCGGCTGCAGGATATGCTTGCTTGGTGTCAGCTTTAGCAAAATTGTATAAAATTAAATCAGATGTCAGTTCAATTGCAAGATTAGGCTCAGGCAGTGCATGTAGAAGTGTTTACGGAGGTTTTGTAAGGTGGCATGCTGGCTCAAAACCTGATGGTAGCGATTCTATAGCCACACAAATTGCAGATTCGAATCATTGGCCTGAAATGCGTGTTTTAGTACTTGTAGTTGGTAATACTCAAAAGAAAGTCAGTTCAACTGTAGGAATGAAAATATCTTCAGAAACATCTGAATTATTAAAACACCGAATACAACACTGTGTTCCCCAAAGAACTGAAGAAATTATAGAAGCTATAAAGAACAAAGACTTTTCTAAATTTGCAGAGATAACAATGAAAGACAGTAATCAGTTTCATGCAATTTGCTTAGATTCATATCCTCCTATTGTGTACATGACAGACATTTCTCATTCAATTGTAGATTTGATTCATAAATATAATGACTTTAGTGGGGTGACAAAAGTAGCATACACATTCGATGCTGGATCGAATGCTTGTTTGTACTTGTTAAAGCATGATGTCCCTAAAGTAATTTCATTAATAAAATATGCATTCCCTTCAACGTCACCAGAAAAATTCATAACAGGTTTAAGTGTTGCCAAAGAAGAAATAAATACTGATATTTTATCAAAGTTGTGTATTGAACCAATGGGAATGGATATGATTAAATACATTATACACACTAAAATAGGTGAAGGACCTACAGAAATAGATTCACATTTGCTGGACAGTAACGGGCTGCCTATTTCAAAGATAAACAAGATATAA
Protein
MSNIVTVIAPVNIAVIKYWGKRDEKLILPLNDSVSATFDTSVMCAKTSVSTHPDFVEDQIWLNGKEESFSNPRLQNCLREIKSRAVAEKTIAEDVLSWKVHVSSENNFPTAAGLASSAAGYACLVSALAKLYKIKSDVSSIARLGSGSACRSVYGGFVRWHAGSKPDGSDSIATQIADSNHWPEMRVLVLVVGNTQKKVSSTVGMKISSETSELLKHRIQHCVPQRTEEIIEAIKNKDFSKFAEITMKDSNQFHAICLDSYPPIVYMTDISHSIVDLIHKYNDFSGVTKVAYTFDAGSNACLYLLKHDVPKVISLIKYAFPSTSPEKFITGLSVAKEEINTDILSKLCIEPMGMDMIKYIIHTKIGEGPTEIDSHLLDSNGLPISKINKI
Summary
Description
Performs the first committed step in the biosynthesis of isoprenes.
Catalytic Activity
(R)-5-diphosphomevalonate + ATP = ADP + CO2 + isopentenyl diphosphate + phosphate
Subunit
Homodimer.
Similarity
Belongs to the diphosphomevalonate decarboxylase family.
Keywords
3D-structure
Acetylation
ATP-binding
Cholesterol biosynthesis
Cholesterol metabolism
Complete proteome
Lipid biosynthesis
Lipid metabolism
Lyase
Nucleotide-binding
Reference proteome
Steroid biosynthesis
Steroid metabolism
Sterol biosynthesis
Sterol metabolism
Feature
chain Diphosphomevalonate decarboxylase
Uniprot
A5A7A2
H9JD62
A0A2A4JDV1
A0A2H1W8I6
A0A2W1BJS3
A0A0C5LBM1
+ More
A0A0L7LJ17 A0A0N0PDF4 A0A194QBH9 A0A212F8V0 S4PE89 A0A2J7RDE7 A0A0A9XIY2 A0A1W4XSS6 A0A1Y1MYD1 I1VX04 A0A023F341 A0A0G3F646 A0A088A0J4 A0A0L7QWY1 A0A1S3JKE6 A0A1B6MMD3 A0A0P4VUR6 R7U748 A0A1S3I4D9 U4TXP9 I1VJ71 A0A224XHX9 J3JYG3 N6TD62 A0A154PAK3 A0A1D2NHI4 A0A1L8DZ38 U5ETY9 A0A1S4F6T1 Q17DS1 A0A1J1J527 T1I721 A0A1L8DZH7 B0W7P4 D6WE42 A0A226DMJ1 A0A087T943 A0A1B0CBF5 E2AFE1 A0A2L2YMZ7 A0A158NDC2 A0A3L8DRS7 A0A3B3Q6V1 A0A131YNJ8 A0A067RMX6 K7ISQ8 A0A131X9U7 A0A224YL36 A0A0H4J596 A0A218USD9 A0A2A3EQS7 A0A3M0K8C1 B4NPC5 A0A336MPX2 C3ZGP5 G3MND8 A0A2U9BCM1 A0A336KUU7 A0A3B3Q5H6 H2VB93 A0A1V4K1D9 A0A3B4Y9W2 A0A3B4VBU7 V9KP19 L7M8G4 A0A1I8NM45 A0A232F494 A0A0C9S2U5 A0A3P8T653 W5K7U2 W5UCK4 W8BAX3 B3NTG5 A0A210PNE5 A0A1I8NCY0 Q3UYC1 Q99JF5 A0A1S3KUY7 A0A060Y7M2 A0A1A6HGM4 G3GRT8 A0A0D2WTJ3 B4KSJ4 F6Y9W8 A0A1W5B150 A0A3B4Z576 H3ABU7 A0A1W4V3D2 B4J7I0 A0A340Y7V9 Q0P570 A0A1S3AAS8 A0A293MV42 A0A1E1X7R0
A0A0L7LJ17 A0A0N0PDF4 A0A194QBH9 A0A212F8V0 S4PE89 A0A2J7RDE7 A0A0A9XIY2 A0A1W4XSS6 A0A1Y1MYD1 I1VX04 A0A023F341 A0A0G3F646 A0A088A0J4 A0A0L7QWY1 A0A1S3JKE6 A0A1B6MMD3 A0A0P4VUR6 R7U748 A0A1S3I4D9 U4TXP9 I1VJ71 A0A224XHX9 J3JYG3 N6TD62 A0A154PAK3 A0A1D2NHI4 A0A1L8DZ38 U5ETY9 A0A1S4F6T1 Q17DS1 A0A1J1J527 T1I721 A0A1L8DZH7 B0W7P4 D6WE42 A0A226DMJ1 A0A087T943 A0A1B0CBF5 E2AFE1 A0A2L2YMZ7 A0A158NDC2 A0A3L8DRS7 A0A3B3Q6V1 A0A131YNJ8 A0A067RMX6 K7ISQ8 A0A131X9U7 A0A224YL36 A0A0H4J596 A0A218USD9 A0A2A3EQS7 A0A3M0K8C1 B4NPC5 A0A336MPX2 C3ZGP5 G3MND8 A0A2U9BCM1 A0A336KUU7 A0A3B3Q5H6 H2VB93 A0A1V4K1D9 A0A3B4Y9W2 A0A3B4VBU7 V9KP19 L7M8G4 A0A1I8NM45 A0A232F494 A0A0C9S2U5 A0A3P8T653 W5K7U2 W5UCK4 W8BAX3 B3NTG5 A0A210PNE5 A0A1I8NCY0 Q3UYC1 Q99JF5 A0A1S3KUY7 A0A060Y7M2 A0A1A6HGM4 G3GRT8 A0A0D2WTJ3 B4KSJ4 F6Y9W8 A0A1W5B150 A0A3B4Z576 H3ABU7 A0A1W4V3D2 B4J7I0 A0A340Y7V9 Q0P570 A0A1S3AAS8 A0A293MV42 A0A1E1X7R0
EC Number
4.1.1.33
Pubmed
17628279
19121390
28756777
26227816
26354079
22118469
+ More
23622113 25401762 26823975 28004739 25474469 27129103 23254933 23537049 22516182 27289101 17510324 18362917 19820115 20798317 26561354 21347285 30249741 29240929 26830274 24845553 20075255 28049606 28797301 26899871 17994087 18563158 22216098 21551351 24402279 25576852 28648823 26131772 25329095 23127152 24495485 28812685 25315136 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 16141072 15489334 24755649 21804562 29704459 17495919 9215903 28503490
23622113 25401762 26823975 28004739 25474469 27129103 23254933 23537049 22516182 27289101 17510324 18362917 19820115 20798317 26561354 21347285 30249741 29240929 26830274 24845553 20075255 28049606 28797301 26899871 17994087 18563158 22216098 21551351 24402279 25576852 28648823 26131772 25329095 23127152 24495485 28812685 25315136 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 16141072 15489334 24755649 21804562 29704459 17495919 9215903 28503490
EMBL
AB274993
BAF62111.1
BABH01036249
BABH01036250
NWSH01001901
PCG69734.1
+ More
PCG69735.1 ODYU01007030 SOQ49415.1 KZ150100 PZC73517.1 KJ951986 AJP70489.1 JTDY01000888 KOB75553.1 KQ460366 KPJ15582.1 KQ459249 KPJ02345.1 AGBW02013108 AGBW02009673 OWR43971.1 OWR50157.1 GAIX01003271 JAA89289.1 NEVH01005291 PNF38856.1 GBHO01024826 GDHC01010516 JAG18778.1 JAQ08113.1 GEZM01017487 JAV90649.1 JQ413986 AFI55102.1 GBBI01003166 JAC15546.1 KP420148 AKJ80168.1 KQ414705 KOC63133.1 GEBQ01002905 JAT37072.1 GDKW01000294 JAI56301.1 AMQN01009085 KB304553 ELU01936.1 KB631811 ERL86359.1 JQ855692 AFI45055.1 GFTR01005772 JAW10654.1 BT128291 AEE63249.1 APGK01042724 KB741007 ENN75638.1 KQ434846 KZC08258.1 LJIJ01000037 ODN04721.1 GFDF01002395 JAV11689.1 GANO01002564 JAB57307.1 CH477291 EAT44593.1 CVRI01000072 CRL07495.1 ACPB03000766 GFDF01002399 JAV11685.1 DS231855 EDS38188.1 KQ971318 EFA00378.2 LNIX01000014 OXA46752.1 KK114061 KFM61632.1 AJWK01005106 AJWK01005107 GL439108 EFN67834.1 IAAA01033033 LAA09486.1 ADTU01012588 QOIP01000005 RLU22873.1 GEDV01009096 JAP79461.1 KK852528 KDR21975.1 AAZX01002897 GEFH01004187 JAP64394.1 GFPF01007212 MAA18358.1 KP689340 AKO63322.1 MUZQ01000150 OWK56674.1 KZ288201 PBC33576.1 QRBI01000116 RMC09342.1 CH964291 EDW86365.1 UFQT01001498 SSX30823.1 GG666620 EEN48271.1 JO843389 AEO35006.1 CP026247 AWP01654.1 UFQS01001020 UFQT01001020 SSX08599.1 SSX28515.1 LSYS01005191 OPJ78246.1 JW867432 AFO99949.1 GACK01004699 JAA60335.1 NNAY01001047 OXU25349.1 GBZX01001376 JAG91364.1 JT411031 AHH39284.1 GAMC01016284 JAB90271.1 CH954180 EDV46969.1 NEDP02005575 OWF37946.1 AK134803 CH466525 BAE22291.1 EDL11681.1 AJ309922 AK089354 AK154883 AK170773 BC008526 FR907970 CDQ87522.1 LZPO01034768 OBS77110.1 JH000004 RAZU01000146 EGV94082.1 RLQ69307.1 KE346368 KJE94983.1 CH933808 EDW09499.2 AFYH01193560 CH916367 EDW01104.1 BC120432 GFWV01019080 MAA43808.1 GFAC01003896 JAT95292.1
PCG69735.1 ODYU01007030 SOQ49415.1 KZ150100 PZC73517.1 KJ951986 AJP70489.1 JTDY01000888 KOB75553.1 KQ460366 KPJ15582.1 KQ459249 KPJ02345.1 AGBW02013108 AGBW02009673 OWR43971.1 OWR50157.1 GAIX01003271 JAA89289.1 NEVH01005291 PNF38856.1 GBHO01024826 GDHC01010516 JAG18778.1 JAQ08113.1 GEZM01017487 JAV90649.1 JQ413986 AFI55102.1 GBBI01003166 JAC15546.1 KP420148 AKJ80168.1 KQ414705 KOC63133.1 GEBQ01002905 JAT37072.1 GDKW01000294 JAI56301.1 AMQN01009085 KB304553 ELU01936.1 KB631811 ERL86359.1 JQ855692 AFI45055.1 GFTR01005772 JAW10654.1 BT128291 AEE63249.1 APGK01042724 KB741007 ENN75638.1 KQ434846 KZC08258.1 LJIJ01000037 ODN04721.1 GFDF01002395 JAV11689.1 GANO01002564 JAB57307.1 CH477291 EAT44593.1 CVRI01000072 CRL07495.1 ACPB03000766 GFDF01002399 JAV11685.1 DS231855 EDS38188.1 KQ971318 EFA00378.2 LNIX01000014 OXA46752.1 KK114061 KFM61632.1 AJWK01005106 AJWK01005107 GL439108 EFN67834.1 IAAA01033033 LAA09486.1 ADTU01012588 QOIP01000005 RLU22873.1 GEDV01009096 JAP79461.1 KK852528 KDR21975.1 AAZX01002897 GEFH01004187 JAP64394.1 GFPF01007212 MAA18358.1 KP689340 AKO63322.1 MUZQ01000150 OWK56674.1 KZ288201 PBC33576.1 QRBI01000116 RMC09342.1 CH964291 EDW86365.1 UFQT01001498 SSX30823.1 GG666620 EEN48271.1 JO843389 AEO35006.1 CP026247 AWP01654.1 UFQS01001020 UFQT01001020 SSX08599.1 SSX28515.1 LSYS01005191 OPJ78246.1 JW867432 AFO99949.1 GACK01004699 JAA60335.1 NNAY01001047 OXU25349.1 GBZX01001376 JAG91364.1 JT411031 AHH39284.1 GAMC01016284 JAB90271.1 CH954180 EDV46969.1 NEDP02005575 OWF37946.1 AK134803 CH466525 BAE22291.1 EDL11681.1 AJ309922 AK089354 AK154883 AK170773 BC008526 FR907970 CDQ87522.1 LZPO01034768 OBS77110.1 JH000004 RAZU01000146 EGV94082.1 RLQ69307.1 KE346368 KJE94983.1 CH933808 EDW09499.2 AFYH01193560 CH916367 EDW01104.1 BC120432 GFWV01019080 MAA43808.1 GFAC01003896 JAT95292.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000235965 UP000192223 UP000005203 UP000053825 UP000085678 UP000014760 UP000030742 UP000019118 UP000076502 UP000094527 UP000008820 UP000183832 UP000015103 UP000002320 UP000007266 UP000198287 UP000054359 UP000092461 UP000000311 UP000005205 UP000279307 UP000261540 UP000027135 UP000002358 UP000197619 UP000242457 UP000269221 UP000007798 UP000001554 UP000246464 UP000005226 UP000190648 UP000261360 UP000261420 UP000095300 UP000215335 UP000265080 UP000018467 UP000221080 UP000008711 UP000242188 UP000095301 UP000000589 UP000087266 UP000193380 UP000092124 UP000001075 UP000273346 UP000008743 UP000009192 UP000002280 UP000192224 UP000261400 UP000008672 UP000192221 UP000001070 UP000265300 UP000009136 UP000079721
UP000235965 UP000192223 UP000005203 UP000053825 UP000085678 UP000014760 UP000030742 UP000019118 UP000076502 UP000094527 UP000008820 UP000183832 UP000015103 UP000002320 UP000007266 UP000198287 UP000054359 UP000092461 UP000000311 UP000005205 UP000279307 UP000261540 UP000027135 UP000002358 UP000197619 UP000242457 UP000269221 UP000007798 UP000001554 UP000246464 UP000005226 UP000190648 UP000261360 UP000261420 UP000095300 UP000215335 UP000265080 UP000018467 UP000221080 UP000008711 UP000242188 UP000095301 UP000000589 UP000087266 UP000193380 UP000092124 UP000001075 UP000273346 UP000008743 UP000009192 UP000002280 UP000192224 UP000261400 UP000008672 UP000192221 UP000001070 UP000265300 UP000009136 UP000079721
Interpro
IPR036554
GHMP_kinase_C_sf
+ More
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006204 GHMP_kinase_N_dom
IPR020568 Ribosomal_S5_D2-typ_fold
IPR005935 Mev_decarb
IPR029765 Mev_diP_decarb
IPR041431 Mvd1_C
IPR005720 Dihydroorotate_DH
IPR027417 P-loop_NTPase
IPR001295 Dihydroorotate_DH_CS
IPR005225 Small_GTP-bd_dom
IPR013785 Aldolase_TIM
IPR001806 Small_GTPase
IPR018935 RIO_kinase_CS
IPR011009 Kinase-like_dom_sf
IPR000687 RIO_kinase
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006204 GHMP_kinase_N_dom
IPR020568 Ribosomal_S5_D2-typ_fold
IPR005935 Mev_decarb
IPR029765 Mev_diP_decarb
IPR041431 Mvd1_C
IPR005720 Dihydroorotate_DH
IPR027417 P-loop_NTPase
IPR001295 Dihydroorotate_DH_CS
IPR005225 Small_GTP-bd_dom
IPR013785 Aldolase_TIM
IPR001806 Small_GTPase
IPR018935 RIO_kinase_CS
IPR011009 Kinase-like_dom_sf
IPR000687 RIO_kinase
Gene 3D
ProteinModelPortal
A5A7A2
H9JD62
A0A2A4JDV1
A0A2H1W8I6
A0A2W1BJS3
A0A0C5LBM1
+ More
A0A0L7LJ17 A0A0N0PDF4 A0A194QBH9 A0A212F8V0 S4PE89 A0A2J7RDE7 A0A0A9XIY2 A0A1W4XSS6 A0A1Y1MYD1 I1VX04 A0A023F341 A0A0G3F646 A0A088A0J4 A0A0L7QWY1 A0A1S3JKE6 A0A1B6MMD3 A0A0P4VUR6 R7U748 A0A1S3I4D9 U4TXP9 I1VJ71 A0A224XHX9 J3JYG3 N6TD62 A0A154PAK3 A0A1D2NHI4 A0A1L8DZ38 U5ETY9 A0A1S4F6T1 Q17DS1 A0A1J1J527 T1I721 A0A1L8DZH7 B0W7P4 D6WE42 A0A226DMJ1 A0A087T943 A0A1B0CBF5 E2AFE1 A0A2L2YMZ7 A0A158NDC2 A0A3L8DRS7 A0A3B3Q6V1 A0A131YNJ8 A0A067RMX6 K7ISQ8 A0A131X9U7 A0A224YL36 A0A0H4J596 A0A218USD9 A0A2A3EQS7 A0A3M0K8C1 B4NPC5 A0A336MPX2 C3ZGP5 G3MND8 A0A2U9BCM1 A0A336KUU7 A0A3B3Q5H6 H2VB93 A0A1V4K1D9 A0A3B4Y9W2 A0A3B4VBU7 V9KP19 L7M8G4 A0A1I8NM45 A0A232F494 A0A0C9S2U5 A0A3P8T653 W5K7U2 W5UCK4 W8BAX3 B3NTG5 A0A210PNE5 A0A1I8NCY0 Q3UYC1 Q99JF5 A0A1S3KUY7 A0A060Y7M2 A0A1A6HGM4 G3GRT8 A0A0D2WTJ3 B4KSJ4 F6Y9W8 A0A1W5B150 A0A3B4Z576 H3ABU7 A0A1W4V3D2 B4J7I0 A0A340Y7V9 Q0P570 A0A1S3AAS8 A0A293MV42 A0A1E1X7R0
A0A0L7LJ17 A0A0N0PDF4 A0A194QBH9 A0A212F8V0 S4PE89 A0A2J7RDE7 A0A0A9XIY2 A0A1W4XSS6 A0A1Y1MYD1 I1VX04 A0A023F341 A0A0G3F646 A0A088A0J4 A0A0L7QWY1 A0A1S3JKE6 A0A1B6MMD3 A0A0P4VUR6 R7U748 A0A1S3I4D9 U4TXP9 I1VJ71 A0A224XHX9 J3JYG3 N6TD62 A0A154PAK3 A0A1D2NHI4 A0A1L8DZ38 U5ETY9 A0A1S4F6T1 Q17DS1 A0A1J1J527 T1I721 A0A1L8DZH7 B0W7P4 D6WE42 A0A226DMJ1 A0A087T943 A0A1B0CBF5 E2AFE1 A0A2L2YMZ7 A0A158NDC2 A0A3L8DRS7 A0A3B3Q6V1 A0A131YNJ8 A0A067RMX6 K7ISQ8 A0A131X9U7 A0A224YL36 A0A0H4J596 A0A218USD9 A0A2A3EQS7 A0A3M0K8C1 B4NPC5 A0A336MPX2 C3ZGP5 G3MND8 A0A2U9BCM1 A0A336KUU7 A0A3B3Q5H6 H2VB93 A0A1V4K1D9 A0A3B4Y9W2 A0A3B4VBU7 V9KP19 L7M8G4 A0A1I8NM45 A0A232F494 A0A0C9S2U5 A0A3P8T653 W5K7U2 W5UCK4 W8BAX3 B3NTG5 A0A210PNE5 A0A1I8NCY0 Q3UYC1 Q99JF5 A0A1S3KUY7 A0A060Y7M2 A0A1A6HGM4 G3GRT8 A0A0D2WTJ3 B4KSJ4 F6Y9W8 A0A1W5B150 A0A3B4Z576 H3ABU7 A0A1W4V3D2 B4J7I0 A0A340Y7V9 Q0P570 A0A1S3AAS8 A0A293MV42 A0A1E1X7R0
PDB
3F0N
E-value=3.06761e-108,
Score=1001
Ontologies
PATHWAY
GO
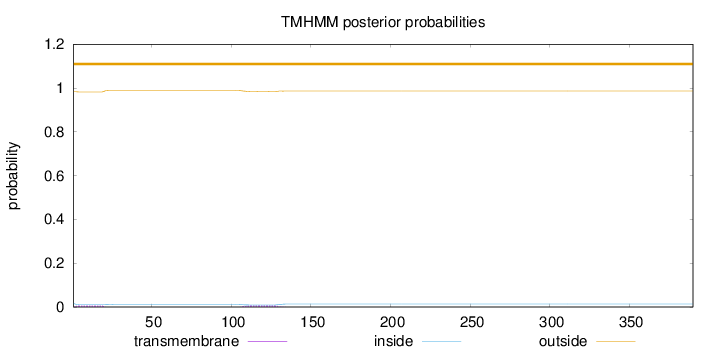
Topology
Length:
390
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28738
Exp number, first 60 AAs:
0.12063
Total prob of N-in:
0.01714
outside
1 - 390
Population Genetic Test Statistics
Pi
307.015168
Theta
242.556715
Tajima's D
0.838543
CLR
0.207299
CSRT
0.607669616519174
Interpretation
Uncertain
