Gene
KWMTBOMO01148 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007477
Annotation
receptor_for_activated_protein_kinase_C_RACK_1_isoform_1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.102
Sequence
CDS
ATGTCTGAAACATTAAAACTCCGAGGAACTCTTAGAGGCCACAATGGCTGGGTTACTCAAATTGCAACTAATCCGAAATACCCGGACATGATCTTATCTTCCTCTCGAGACAAAACTCTCATCGTGTGGAAGCTGACCAGAGACGAAAACAACTACGGTATACCTCAAAAGCGTTTATACGGTCATTCGCACTTCATTTCTGATGTTGTGCTGTCTAGTGACGGTAATTACGCCCTTTCCGGTTCTTGGGACAAGACTTTGCGTTTGTGGGATCTCGCTGCAGGCAAGACTACCAGACGATTCGAGGACCATACTAAGGATGTACTCTCGGTAGCCTTCTCAGTTGACAACCGTCAGATAGTGTCTGGTTCTCGCGACAAGACTATCAAACTCTGGAACACCCTTGCGGAGTGCAAGTATACCATCCAAGATGATGGACACAGCGATTGGGTGTCATGTGTCAGATTCTCACCCAATCATGCCAACCCCATTATTGTATCCTGTGGTTGGGACAGAACTGTCAAGGTCTGGCATCTCACTAACTGTAAGCTCAAGATTAACCATTTGGGTCACTCTGGCTATCTGAACACTGTTACTGTATCACCTGATGGCTCTTTGTGTGCATCTGGTGGTAAGGATATGAAGGCGATGTTGTGGGATCTGAACGATGGCAAACACCTCCACACCTTAGACCACAATGATATCATCACGGCCTTGTGCTTCTCACCCAACAGATACTGGCTATGTGCTGCTTTCGGACCTTCCATCAAGATCTGGGATCTGGAAAGCAAGGAGATGGTTGAAGAGCTCAGGCCTGAAATCATTAACCAAACGCAAACTTCCAAGACAGACCCTCCTCAGTGTCTCTCCCTAGCATGGTCTACTGATGGACAAACCTTATTTGCTGGTTACTCAGATAATACTATAAGAGTGTGGCAGGTTTCCATCTCAGCTCGATAA
Protein
MSETLKLRGTLRGHNGWVTQIATNPKYPDMILSSSRDKTLIVWKLTRDENNYGIPQKRLYGHSHFISDVVLSSDGNYALSGSWDKTLRLWDLAAGKTTRRFEDHTKDVLSVAFSVDNRQIVSGSRDKTIKLWNTLAECKYTIQDDGHSDWVSCVRFSPNHANPIIVSCGWDRTVKVWHLTNCKLKINHLGHSGYLNTVTVSPDGSLCASGGKDMKAMLWDLNDGKHLHTLDHNDIITALCFSPNRYWLCAAFGPSIKIWDLESKEMVEELRPEIINQTQTSKTDPPQCLSLAWSTDGQTLFAGYSDNTIRVWQVSISAR
Summary
Uniprot
Q2F5M4
H9JD80
C4PG27
Q60FS2
A0A2A4JEA3
A0A2H1WAA4
+ More
F5BYI2 A0A3G1T1C1 A9LST1 Q2FBK6 I4DM37 A0A0N0PD51 I4DIH7 E3UKN0 A0A0L7LSA8 Q2FBK5 F8UZU8 Q86SB4 Q95PD5 S4PX02 A0A2J7QGP3 A0A067RGF2 Q09KA1 A0A0J7L0K1 A0A195DI46 N6SU69 H9LD14 J3JX55 D6WUS7 A0A154NVR6 A0A023F933 E1ZX95 A0A2A3E9V2 A0A088A2A0 V9IF58 A0A026WLF9 A0A224XU06 K7IXB4 A0A069DSH4 A0A0A9VPY7 A0A1Y1MBB8 E2B2K1 A0A0P4VYB2 A0A0T6AVD5 R4G4R2 A0A1W4WU03 A0A212F8S9 A0A1B6FWW0 E9IG50 F4WBU4 A0A195BD72 A0A195FS91 A0A195C2Z0 A0A310SFD9 A0A151WNA4 A0A1B6DLQ3 A0A0V0G2L4 A0A1D1YVS1 R4WI41 A0A0L7R3Q8 A0A2H8TWZ2 Q5XUC3 J9JV14 C4WT57 U5EMN4 Q6PPH2 A0A1B6HCE2 A0A1L8DMF0 E0VKT1 A0A0N8ECC5 A2I3W1 A0A1I9WLK9 A0A1S3D6L1 G8CV13 A0A0P6IWS7 A0A0K8TQI5 A0A1Q3G286 B0X0Y1 Q1HRQ2 A0A023EN53 A0A1J1I321 A0A2R7WYN8 A0A034W366 A0A0A1X351 W8AZB4 C6ZQW2 A0A182P2B8 A0A1I8QBP5 A0A1L8EF41 B4LSR9 A0A182V9W1 A0A182U6S6 A0A182KJN0 A0A182XB19 Q7Q0T5 A0A182I0L5 A0A182ITH4 A0A1L8EEZ0 T1PFF5 A0A182MAF1
F5BYI2 A0A3G1T1C1 A9LST1 Q2FBK6 I4DM37 A0A0N0PD51 I4DIH7 E3UKN0 A0A0L7LSA8 Q2FBK5 F8UZU8 Q86SB4 Q95PD5 S4PX02 A0A2J7QGP3 A0A067RGF2 Q09KA1 A0A0J7L0K1 A0A195DI46 N6SU69 H9LD14 J3JX55 D6WUS7 A0A154NVR6 A0A023F933 E1ZX95 A0A2A3E9V2 A0A088A2A0 V9IF58 A0A026WLF9 A0A224XU06 K7IXB4 A0A069DSH4 A0A0A9VPY7 A0A1Y1MBB8 E2B2K1 A0A0P4VYB2 A0A0T6AVD5 R4G4R2 A0A1W4WU03 A0A212F8S9 A0A1B6FWW0 E9IG50 F4WBU4 A0A195BD72 A0A195FS91 A0A195C2Z0 A0A310SFD9 A0A151WNA4 A0A1B6DLQ3 A0A0V0G2L4 A0A1D1YVS1 R4WI41 A0A0L7R3Q8 A0A2H8TWZ2 Q5XUC3 J9JV14 C4WT57 U5EMN4 Q6PPH2 A0A1B6HCE2 A0A1L8DMF0 E0VKT1 A0A0N8ECC5 A2I3W1 A0A1I9WLK9 A0A1S3D6L1 G8CV13 A0A0P6IWS7 A0A0K8TQI5 A0A1Q3G286 B0X0Y1 Q1HRQ2 A0A023EN53 A0A1J1I321 A0A2R7WYN8 A0A034W366 A0A0A1X351 W8AZB4 C6ZQW2 A0A182P2B8 A0A1I8QBP5 A0A1L8EF41 B4LSR9 A0A182V9W1 A0A182U6S6 A0A182KJN0 A0A182XB19 Q7Q0T5 A0A182I0L5 A0A182ITH4 A0A1L8EEZ0 T1PFF5 A0A182MAF1
Pubmed
19121390
21452889
28756777
16469071
22651552
26354079
+ More
26227816 23622113 24845553 23537049 22516182 18362917 19820115 25474469 20798317 24508170 30249741 20075255 26334808 25401762 26823975 28004739 27129103 22118469 21282665 21719571 23691247 20566863 27538518 22028913 29136191 26369729 17204158 17510324 24945155 25348373 25830018 24495485 20496585 17994087 20966253 12364791 14747013 17210077 25315136
26227816 23622113 24845553 23537049 22516182 18362917 19820115 25474469 20798317 24508170 30249741 20075255 26334808 25401762 26823975 28004739 27129103 22118469 21282665 21719571 23691247 20566863 27538518 22028913 29136191 26369729 17204158 17510324 24945155 25348373 25830018 24495485 20496585 17994087 20966253 12364791 14747013 17210077 25315136
EMBL
DQ311399
HQ638203
ABD36343.1
AEH16566.1
BABH01036249
FJ943495
+ More
ACR19030.1 AB189027 BAD52259.1 NWSH01001901 PCG69733.1 ODYU01007030 SOQ49414.1 JF417987 KZ150100 AEB26318.1 PZC73518.1 MG846923 AXY94775.1 EU259815 ABX55886.1 DQ073455 AAZ29605.1 AK402355 BAM18977.1 KQ460366 KPJ15581.1 AK401095 KQ459249 BAM17717.1 KPJ02344.1 HM449904 ADO33039.1 JTDY01000193 KOB78365.1 DQ073456 AAZ29606.1 JF979035 AEI84399.1 AB089281 BAC56715.1 AF368031 AAK51552.1 GAIX01005893 JAA86667.1 NEVH01014361 PNF27748.1 KK852699 KDR18184.1 DQ885470 ABI63548.1 LBMM01001608 KMQ96024.1 KQ980824 KYN12568.1 APGK01056999 APGK01057000 KB741277 ENN71239.1 HM130685 AEA72283.1 BT127823 AEE62785.1 KQ971363 EFA09061.1 KQ434769 KZC03785.1 GBBI01000720 GEMB01001668 JAC17992.1 JAS01495.1 GL435030 EFN74185.1 KZ288311 PBC28507.1 JR040024 JR040025 AEY58953.1 AEY58954.1 KK107154 QOIP01000001 EZA56887.1 RLU27536.1 GFTR01004967 JAW11459.1 AAZX01011471 GBGD01002203 JAC86686.1 GBHO01045067 GBRD01006980 GDHC01003293 JAF98536.1 JAG58841.1 JAQ15336.1 GEZM01036964 JAV82308.1 GL445147 EFN90078.1 GDKW01000983 JAI55612.1 LJIG01022720 KRT79066.1 ACPB03007814 GAHY01000561 JAA76949.1 AGBW02009673 OWR50156.1 GECZ01015246 JAS54523.1 GL762910 EFZ20464.1 GL888066 EGI68372.1 KQ976511 KYM82521.1 KQ981280 KYN43455.1 KQ978317 KYM95229.1 KQ761976 OAD56421.1 KQ982911 KYQ49323.1 GEDC01010684 JAS26614.1 GECL01003775 JAP02349.1 GDJX01009214 JAT58722.1 AK417015 BAN20230.1 KQ414663 KOC65376.1 GFXV01006951 MBW18756.1 AY737531 AAU84924.1 ABLF02035580 AK340474 BAH71077.1 GANO01000944 JAB58927.1 AY588074 AAT01086.1 GECU01035312 JAS72394.1 GFDF01006537 JAV07547.1 DS235250 EEB13987.1 GDIQ01040668 JAN54069.1 EF070464 ABM55530.1 KU932393 APA34029.1 HQ385972 KT724285 QKKF02022802 ADQ73873.1 AMK48127.1 RZF38185.1 GDIQ01003331 JAN91406.1 GDAI01001448 JAI16155.1 GFDL01001133 JAV33912.1 DS232248 EDS38373.1 DQ440042 CH477971 ABF18075.1 EAT34720.1 GAPW01002770 GAPW01002769 JAC10828.1 CVRI01000038 CRK94178.1 KK855891 PTY24280.1 GAKP01010382 JAC48570.1 GBXI01009016 JAD05276.1 GAMC01016362 JAB90193.1 EZ114913 ACU30966.1 GFDG01001544 JAV17255.1 CH940649 EDW63808.1 AAAB01008980 EAA13872.2 APCN01005219 GFDG01001565 JAV17234.1 KA647419 AFP62048.1 AXCM01000013
ACR19030.1 AB189027 BAD52259.1 NWSH01001901 PCG69733.1 ODYU01007030 SOQ49414.1 JF417987 KZ150100 AEB26318.1 PZC73518.1 MG846923 AXY94775.1 EU259815 ABX55886.1 DQ073455 AAZ29605.1 AK402355 BAM18977.1 KQ460366 KPJ15581.1 AK401095 KQ459249 BAM17717.1 KPJ02344.1 HM449904 ADO33039.1 JTDY01000193 KOB78365.1 DQ073456 AAZ29606.1 JF979035 AEI84399.1 AB089281 BAC56715.1 AF368031 AAK51552.1 GAIX01005893 JAA86667.1 NEVH01014361 PNF27748.1 KK852699 KDR18184.1 DQ885470 ABI63548.1 LBMM01001608 KMQ96024.1 KQ980824 KYN12568.1 APGK01056999 APGK01057000 KB741277 ENN71239.1 HM130685 AEA72283.1 BT127823 AEE62785.1 KQ971363 EFA09061.1 KQ434769 KZC03785.1 GBBI01000720 GEMB01001668 JAC17992.1 JAS01495.1 GL435030 EFN74185.1 KZ288311 PBC28507.1 JR040024 JR040025 AEY58953.1 AEY58954.1 KK107154 QOIP01000001 EZA56887.1 RLU27536.1 GFTR01004967 JAW11459.1 AAZX01011471 GBGD01002203 JAC86686.1 GBHO01045067 GBRD01006980 GDHC01003293 JAF98536.1 JAG58841.1 JAQ15336.1 GEZM01036964 JAV82308.1 GL445147 EFN90078.1 GDKW01000983 JAI55612.1 LJIG01022720 KRT79066.1 ACPB03007814 GAHY01000561 JAA76949.1 AGBW02009673 OWR50156.1 GECZ01015246 JAS54523.1 GL762910 EFZ20464.1 GL888066 EGI68372.1 KQ976511 KYM82521.1 KQ981280 KYN43455.1 KQ978317 KYM95229.1 KQ761976 OAD56421.1 KQ982911 KYQ49323.1 GEDC01010684 JAS26614.1 GECL01003775 JAP02349.1 GDJX01009214 JAT58722.1 AK417015 BAN20230.1 KQ414663 KOC65376.1 GFXV01006951 MBW18756.1 AY737531 AAU84924.1 ABLF02035580 AK340474 BAH71077.1 GANO01000944 JAB58927.1 AY588074 AAT01086.1 GECU01035312 JAS72394.1 GFDF01006537 JAV07547.1 DS235250 EEB13987.1 GDIQ01040668 JAN54069.1 EF070464 ABM55530.1 KU932393 APA34029.1 HQ385972 KT724285 QKKF02022802 ADQ73873.1 AMK48127.1 RZF38185.1 GDIQ01003331 JAN91406.1 GDAI01001448 JAI16155.1 GFDL01001133 JAV33912.1 DS232248 EDS38373.1 DQ440042 CH477971 ABF18075.1 EAT34720.1 GAPW01002770 GAPW01002769 JAC10828.1 CVRI01000038 CRK94178.1 KK855891 PTY24280.1 GAKP01010382 JAC48570.1 GBXI01009016 JAD05276.1 GAMC01016362 JAB90193.1 EZ114913 ACU30966.1 GFDG01001544 JAV17255.1 CH940649 EDW63808.1 AAAB01008980 EAA13872.2 APCN01005219 GFDG01001565 JAV17234.1 KA647419 AFP62048.1 AXCM01000013
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000235965
+ More
UP000027135 UP000036403 UP000078492 UP000019118 UP000007266 UP000076502 UP000000311 UP000242457 UP000005203 UP000053097 UP000279307 UP000002358 UP000008237 UP000015103 UP000192223 UP000007151 UP000007755 UP000078540 UP000078541 UP000078542 UP000075809 UP000053825 UP000007819 UP000009046 UP000079169 UP000291343 UP000002320 UP000008820 UP000183832 UP000075885 UP000095300 UP000008792 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075880 UP000095301 UP000075883
UP000027135 UP000036403 UP000078492 UP000019118 UP000007266 UP000076502 UP000000311 UP000242457 UP000005203 UP000053097 UP000279307 UP000002358 UP000008237 UP000015103 UP000192223 UP000007151 UP000007755 UP000078540 UP000078541 UP000078542 UP000075809 UP000053825 UP000007819 UP000009046 UP000079169 UP000291343 UP000002320 UP000008820 UP000183832 UP000075885 UP000095300 UP000008792 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075880 UP000095301 UP000075883
PRIDE
Pfam
PF00400 WD40
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
Q2F5M4
H9JD80
C4PG27
Q60FS2
A0A2A4JEA3
A0A2H1WAA4
+ More
F5BYI2 A0A3G1T1C1 A9LST1 Q2FBK6 I4DM37 A0A0N0PD51 I4DIH7 E3UKN0 A0A0L7LSA8 Q2FBK5 F8UZU8 Q86SB4 Q95PD5 S4PX02 A0A2J7QGP3 A0A067RGF2 Q09KA1 A0A0J7L0K1 A0A195DI46 N6SU69 H9LD14 J3JX55 D6WUS7 A0A154NVR6 A0A023F933 E1ZX95 A0A2A3E9V2 A0A088A2A0 V9IF58 A0A026WLF9 A0A224XU06 K7IXB4 A0A069DSH4 A0A0A9VPY7 A0A1Y1MBB8 E2B2K1 A0A0P4VYB2 A0A0T6AVD5 R4G4R2 A0A1W4WU03 A0A212F8S9 A0A1B6FWW0 E9IG50 F4WBU4 A0A195BD72 A0A195FS91 A0A195C2Z0 A0A310SFD9 A0A151WNA4 A0A1B6DLQ3 A0A0V0G2L4 A0A1D1YVS1 R4WI41 A0A0L7R3Q8 A0A2H8TWZ2 Q5XUC3 J9JV14 C4WT57 U5EMN4 Q6PPH2 A0A1B6HCE2 A0A1L8DMF0 E0VKT1 A0A0N8ECC5 A2I3W1 A0A1I9WLK9 A0A1S3D6L1 G8CV13 A0A0P6IWS7 A0A0K8TQI5 A0A1Q3G286 B0X0Y1 Q1HRQ2 A0A023EN53 A0A1J1I321 A0A2R7WYN8 A0A034W366 A0A0A1X351 W8AZB4 C6ZQW2 A0A182P2B8 A0A1I8QBP5 A0A1L8EF41 B4LSR9 A0A182V9W1 A0A182U6S6 A0A182KJN0 A0A182XB19 Q7Q0T5 A0A182I0L5 A0A182ITH4 A0A1L8EEZ0 T1PFF5 A0A182MAF1
F5BYI2 A0A3G1T1C1 A9LST1 Q2FBK6 I4DM37 A0A0N0PD51 I4DIH7 E3UKN0 A0A0L7LSA8 Q2FBK5 F8UZU8 Q86SB4 Q95PD5 S4PX02 A0A2J7QGP3 A0A067RGF2 Q09KA1 A0A0J7L0K1 A0A195DI46 N6SU69 H9LD14 J3JX55 D6WUS7 A0A154NVR6 A0A023F933 E1ZX95 A0A2A3E9V2 A0A088A2A0 V9IF58 A0A026WLF9 A0A224XU06 K7IXB4 A0A069DSH4 A0A0A9VPY7 A0A1Y1MBB8 E2B2K1 A0A0P4VYB2 A0A0T6AVD5 R4G4R2 A0A1W4WU03 A0A212F8S9 A0A1B6FWW0 E9IG50 F4WBU4 A0A195BD72 A0A195FS91 A0A195C2Z0 A0A310SFD9 A0A151WNA4 A0A1B6DLQ3 A0A0V0G2L4 A0A1D1YVS1 R4WI41 A0A0L7R3Q8 A0A2H8TWZ2 Q5XUC3 J9JV14 C4WT57 U5EMN4 Q6PPH2 A0A1B6HCE2 A0A1L8DMF0 E0VKT1 A0A0N8ECC5 A2I3W1 A0A1I9WLK9 A0A1S3D6L1 G8CV13 A0A0P6IWS7 A0A0K8TQI5 A0A1Q3G286 B0X0Y1 Q1HRQ2 A0A023EN53 A0A1J1I321 A0A2R7WYN8 A0A034W366 A0A0A1X351 W8AZB4 C6ZQW2 A0A182P2B8 A0A1I8QBP5 A0A1L8EF41 B4LSR9 A0A182V9W1 A0A182U6S6 A0A182KJN0 A0A182XB19 Q7Q0T5 A0A182I0L5 A0A182ITH4 A0A1L8EEZ0 T1PFF5 A0A182MAF1
PDB
4V6W
E-value=3.50788e-157,
Score=1422
Ontologies
GO
GO:0016301
GO:0005829
GO:0005634
GO:0005080
GO:0043022
GO:0019899
GO:0001934
GO:0072344
GO:0022627
GO:0003743
GO:0004402
GO:0001933
GO:0044829
GO:0009267
GO:0075522
GO:0048477
GO:0046716
GO:0016243
GO:0030838
GO:0035220
GO:0007626
GO:0005776
GO:0042335
GO:0018991
GO:0045725
GO:0005515
GO:0016021
GO:0016020
GO:0022891
GO:0022857
GO:0045905
GO:0006334
GO:0003779
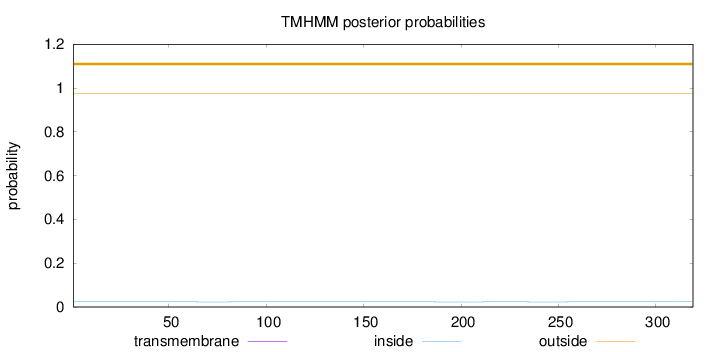
Topology
Length:
319
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00191
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.02343
outside
1 - 319
Population Genetic Test Statistics
Pi
167.571826
Theta
159.714579
Tajima's D
-0.43206
CLR
0.2019
CSRT
0.257937103144843
Interpretation
Uncertain
