Gene
KWMTBOMO01143
Pre Gene Modal
BGIBMGA007462
Annotation
PREDICTED:_sialin_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.967
Sequence
CDS
ATGGACGTGCCGGCAAAGGGAAATGTTCTGGGACGTGTCGTCCCAGCACGCTATGTCCTCGCCGTCCTTGGCTCCATTGGCATGGCCATAGTCTACGGGTTGAAGGTCAATCTGTCGGTGGCGCTAGTGGGGATGCTGAACAAAACAGCGATCAAAATAGCTCTGGGCAATAAGAGTCATGACGAAACTTTATTTAAGAACCTGTCAGTGATACCCGACGATGAGGAATGCCAGCCAACTGGTGGATCTAATAGAAACGCTACGGAATTGGTGGGACCGTTCGTTTGGTCATCCGAAGTGCAGGGAATCGTTTTGAGTTGCTACTTCTGGGGTTATTTCGTCTCCCAAATACCTGGGGCTAGAGTAGCAGAACTTTTCTCGGCGAAATGGGTGATGTTCTTCAGTGTAGCCATCAACGTGGTTTGCACATTGCTCACCCCGGTGATGGCAGAGGCACATTATATTGCCGTTGTAATCATGCGAGTCGGAGAAGGCATAGGTGGGGGAGTAACATTTCCAGCGATGCATGTCTTGTTATCAAAATGGGCGCCTCCAGCTGAACGAAGTGTCATGGCGGCGTTGGTTTACGCGGGAACTTCTCTTGGTACTGTCATTTCTATGTTGATGGCTGGCGTTCTTACTGCTAATGTTGGTTGGGAGAGTGTTTTCTACGTGATGGGTGGTCTGTCGTGCGTCTGGTGTGTGCTTTGGGTGATCTTAGTCCAGGACTCACCCCAGCAACAGCCACTTATCAGCGCGGAAGAGAGAAGCATGATCGTGAGCTCGCTGGGCGGAAAAACTAATGACAAATCCCATAAGAAATTACCTGTACCTTGGAGTAAGGTGCTCAAGTCACCCCCGTTCCTGGCGATCTTAGTGTCGCACGCGTGCTCCAACTGGGGCTGGTACATGTTACTTATCGAGACACCTTTCTATATGGACCAAGTACTCAAGTTCGACATGACCGAGAATGCGGTCATAACTGCTCTCCCGTTCCTTTCCCTGTGGATTTTCAGTATTGCTCTGAGCAGAACCTTAGATTGGCTTCGTTCTAAGAACATTATCACAACTACTACAGCTAGAAAGATTGGCACTCTGTTCGCTTCACTGGTACCGGCAGCCTGTCTACTGGGGCTATGCTACATCGGGTGTAACCGCGCTGGCGCCGTGGTGCTGATGGCCGTGGGCATCACCAGCATCGGCGGAATGTTCTGCGGATTCCTTTCTAATCACATCGACATTGCTCCTAACTTTGCCGGCACCCTGATGGCGATGACAAACACGGTGGCTACCATCCCGGGAATTGTGGTCCCCATTTTTGTTGGAATCCTGACCCACGGGAATCAAACTATTTCGGCTTGGCGTGTCATATTCTTCGTGACAATTGGACTCTACATTATTGAAATTATAGCGTACACTTTATTCGGTTCTGGGGAAGAGCAGTCTTGGAACAAAGTGACAGAAAATAAAGAGGACTTTGAGGTGAAGCCTCTCAAGAACGAAAAGACGCACGTCTAA
Protein
MDVPAKGNVLGRVVPARYVLAVLGSIGMAIVYGLKVNLSVALVGMLNKTAIKIALGNKSHDETLFKNLSVIPDDEECQPTGGSNRNATELVGPFVWSSEVQGIVLSCYFWGYFVSQIPGARVAELFSAKWVMFFSVAINVVCTLLTPVMAEAHYIAVVIMRVGEGIGGGVTFPAMHVLLSKWAPPAERSVMAALVYAGTSLGTVISMLMAGVLTANVGWESVFYVMGGLSCVWCVLWVILVQDSPQQQPLISAEERSMIVSSLGGKTNDKSHKKLPVPWSKVLKSPPFLAILVSHACSNWGWYMLLIETPFYMDQVLKFDMTENAVITALPFLSLWIFSIALSRTLDWLRSKNIITTTTARKIGTLFASLVPAACLLGLCYIGCNRAGAVVLMAVGITSIGGMFCGFLSNHIDIAPNFAGTLMAMTNTVATIPGIVVPIFVGILTHGNQTISAWRVIFFVTIGLYIIEIIAYTLFGSGEEQSWNKVTENKEDFEVKPLKNEKTHV
Summary
Uniprot
H9JD65
A0A194QFW7
A0A1S4FWV3
A0A336LH25
A0A212FIW4
A0A182URL5
+ More
A0A182L660 Q7Q1S5 A0A182ICS7 A0A084VWB7 Q16LU3 A0A182RGQ2 A0A182QU08 A0A182VTW5 A0A182PKV1 A0A182JFI1 A0A1Q3FRX6 A0A182JNR7 A0A1B0DIM0 A0A182Y8B4 A0A182WZE0 A0A182TN75 A0A182NAX9 A0A182F4D4 A0A1I8QB07 B0WCJ3 A0A182MCR0 A0A182GCT3 A0A0L0CI44 A0A1I8N9U6 A0A1A9UTW9 A0A1B0FMP5 A0A1A9Z5X8 A0A1B0BDS9 V5GWS5 A0A1Q3FRW5 B4KEU3 B4JDF0 W5J4J0 B4LU37 Q29LH5 B4GPY3 A0A0M4EEJ2 A0A1B0CDU0 A0A1A9WUC1 A0A1A9XYT0 B4MZJ1 B3N8C2 B4P311 B3MN94 A0A1W4VYB8 A0A3B0JC14 Q9VPX2 B4ID58 W8BWB5 A0A0J9QTR8 A0A0A1WZ82 A0A034WUJ8 T1DKQ2 A0A088A6U6 A0A154PL53 A0A0L7QLU6 A0A310SFK0 E2AHU1 A0A0N0U7D6 T1PBF5 A0A151IAL0 A0A158NEF3 F4WVK5 A0A195BV40 A0A1Y1NGT2 A0A195FL16 E2BIX9 A0A195EP60 A0A0J7KZ16 A0A0T6AZQ5 A0A2A3E9L3 K7IRU9 E9GHP0 A0A0P5ZVD2 A0A232EHZ9 A0A162D4Z8 A0A0P5QI90 A0A0P4ZW54 A0A0P5KF83 A0A0P6IKX3 A0A0P5G067 A0A0P5GR19 B4KEU4 E9GHN7 E9GKW4 E9GHN9 A0A0K8W7Z8 A0A0P5LAA3 A0A0P4WPA4 A0A0P5YKS6 A0A1J1HJ49 A0A0P6BRB5 A0A0P5KMP1 A0A0T6AUS9
A0A182L660 Q7Q1S5 A0A182ICS7 A0A084VWB7 Q16LU3 A0A182RGQ2 A0A182QU08 A0A182VTW5 A0A182PKV1 A0A182JFI1 A0A1Q3FRX6 A0A182JNR7 A0A1B0DIM0 A0A182Y8B4 A0A182WZE0 A0A182TN75 A0A182NAX9 A0A182F4D4 A0A1I8QB07 B0WCJ3 A0A182MCR0 A0A182GCT3 A0A0L0CI44 A0A1I8N9U6 A0A1A9UTW9 A0A1B0FMP5 A0A1A9Z5X8 A0A1B0BDS9 V5GWS5 A0A1Q3FRW5 B4KEU3 B4JDF0 W5J4J0 B4LU37 Q29LH5 B4GPY3 A0A0M4EEJ2 A0A1B0CDU0 A0A1A9WUC1 A0A1A9XYT0 B4MZJ1 B3N8C2 B4P311 B3MN94 A0A1W4VYB8 A0A3B0JC14 Q9VPX2 B4ID58 W8BWB5 A0A0J9QTR8 A0A0A1WZ82 A0A034WUJ8 T1DKQ2 A0A088A6U6 A0A154PL53 A0A0L7QLU6 A0A310SFK0 E2AHU1 A0A0N0U7D6 T1PBF5 A0A151IAL0 A0A158NEF3 F4WVK5 A0A195BV40 A0A1Y1NGT2 A0A195FL16 E2BIX9 A0A195EP60 A0A0J7KZ16 A0A0T6AZQ5 A0A2A3E9L3 K7IRU9 E9GHP0 A0A0P5ZVD2 A0A232EHZ9 A0A162D4Z8 A0A0P5QI90 A0A0P4ZW54 A0A0P5KF83 A0A0P6IKX3 A0A0P5G067 A0A0P5GR19 B4KEU4 E9GHN7 E9GKW4 E9GHN9 A0A0K8W7Z8 A0A0P5LAA3 A0A0P4WPA4 A0A0P5YKS6 A0A1J1HJ49 A0A0P6BRB5 A0A0P5KMP1 A0A0T6AUS9
Pubmed
19121390
26354079
22118469
20966253
12364791
14747013
+ More
17210077 24438588 17510324 25244985 26483478 26108605 25315136 17994087 20920257 23761445 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 22936249 25830018 25348373 20798317 21347285 21719571 28004739 20075255 21292972 28648823
17210077 24438588 17510324 25244985 26483478 26108605 25315136 17994087 20920257 23761445 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 22936249 25830018 25348373 20798317 21347285 21719571 28004739 20075255 21292972 28648823
EMBL
BABH01036245
KQ459249
KPJ02341.1
UFQS01003761
UFQT01003761
SSX15915.1
+ More
SSX35257.1 AGBW02008325 OWR53660.1 AAAB01008980 EAA13886.3 APCN01005756 ATLV01017545 ATLV01017546 KE525174 KFB42261.1 CH477890 EAT35306.1 AXCN02000652 GFDL01004740 JAV30305.1 AJVK01006146 DS231888 EDS43564.1 AXCM01001829 JXUM01054802 JXUM01054803 KQ561837 KXJ77387.1 JRES01000438 KNC31174.1 CCAG010018215 JXJN01012662 GALX01002289 JAB66177.1 GFDL01004721 JAV30324.1 CH933807 EDW12993.1 CH916368 EDW03320.1 ADMH02002138 ETN58333.1 CH940649 EDW65090.1 CH379060 EAL34070.1 CH479187 EDW39655.1 CP012523 ALC38580.1 AJWK01008230 AJWK01008231 AJWK01008232 AJWK01008233 CH963920 EDW77776.1 CH954177 EDV57309.1 CM000157 EDW87218.1 CH902620 EDV31051.2 OUUW01000004 SPP79917.1 AE014134 AY095062 AAF51413.1 AAM11390.1 CH480829 EDW45484.1 GAMC01000945 JAC05611.1 CM002910 KMY87426.1 GBXI01010507 JAD03785.1 GAKP01001489 JAC57463.1 GAMD01000846 JAB00745.1 KQ434936 KZC12018.1 KQ414905 KOC59597.1 KQ776753 OAD52180.1 GL439579 EFN67020.1 KQ435709 KOX80058.1 KA646096 AFP60725.1 KQ978221 KYM96280.1 ADTU01013276 GL888387 EGI61826.1 KQ976401 KYM92489.1 GEZM01002874 GEZM01002873 GEZM01002872 GEZM01002871 JAV97102.1 KQ981490 KYN41108.1 GL448543 EFN84324.1 KQ978625 KYN29677.1 LBMM01001905 KMQ95571.1 LJIG01022482 KRT80341.1 KZ288312 PBC28415.1 GL732545 EFX81019.1 GDIP01038502 JAM65213.1 NNAY01004388 OXU17964.1 LRGB01002580 KZS06984.1 GDIQ01120286 JAL31440.1 GDIP01207930 JAJ15472.1 GDIQ01190370 GDIQ01190369 GDIQ01108849 GDIQ01065966 JAK61356.1 GDIQ01002786 JAN91951.1 GDIQ01269438 JAJ82286.1 GDIQ01238847 JAK12878.1 EDW12994.1 EFX81022.1 GL732550 EFX79896.1 EFX81020.1 GDHF01005140 JAI47174.1 GDIQ01172818 JAK78907.1 GDRN01074321 JAI63263.1 GDIP01056408 JAM47307.1 CVRI01000001 CRK86494.1 GDIP01012441 JAM91274.1 GDIQ01189412 JAK62313.1 LJIG01022770 KRT78806.1
SSX35257.1 AGBW02008325 OWR53660.1 AAAB01008980 EAA13886.3 APCN01005756 ATLV01017545 ATLV01017546 KE525174 KFB42261.1 CH477890 EAT35306.1 AXCN02000652 GFDL01004740 JAV30305.1 AJVK01006146 DS231888 EDS43564.1 AXCM01001829 JXUM01054802 JXUM01054803 KQ561837 KXJ77387.1 JRES01000438 KNC31174.1 CCAG010018215 JXJN01012662 GALX01002289 JAB66177.1 GFDL01004721 JAV30324.1 CH933807 EDW12993.1 CH916368 EDW03320.1 ADMH02002138 ETN58333.1 CH940649 EDW65090.1 CH379060 EAL34070.1 CH479187 EDW39655.1 CP012523 ALC38580.1 AJWK01008230 AJWK01008231 AJWK01008232 AJWK01008233 CH963920 EDW77776.1 CH954177 EDV57309.1 CM000157 EDW87218.1 CH902620 EDV31051.2 OUUW01000004 SPP79917.1 AE014134 AY095062 AAF51413.1 AAM11390.1 CH480829 EDW45484.1 GAMC01000945 JAC05611.1 CM002910 KMY87426.1 GBXI01010507 JAD03785.1 GAKP01001489 JAC57463.1 GAMD01000846 JAB00745.1 KQ434936 KZC12018.1 KQ414905 KOC59597.1 KQ776753 OAD52180.1 GL439579 EFN67020.1 KQ435709 KOX80058.1 KA646096 AFP60725.1 KQ978221 KYM96280.1 ADTU01013276 GL888387 EGI61826.1 KQ976401 KYM92489.1 GEZM01002874 GEZM01002873 GEZM01002872 GEZM01002871 JAV97102.1 KQ981490 KYN41108.1 GL448543 EFN84324.1 KQ978625 KYN29677.1 LBMM01001905 KMQ95571.1 LJIG01022482 KRT80341.1 KZ288312 PBC28415.1 GL732545 EFX81019.1 GDIP01038502 JAM65213.1 NNAY01004388 OXU17964.1 LRGB01002580 KZS06984.1 GDIQ01120286 JAL31440.1 GDIP01207930 JAJ15472.1 GDIQ01190370 GDIQ01190369 GDIQ01108849 GDIQ01065966 JAK61356.1 GDIQ01002786 JAN91951.1 GDIQ01269438 JAJ82286.1 GDIQ01238847 JAK12878.1 EDW12994.1 EFX81022.1 GL732550 EFX79896.1 EFX81020.1 GDHF01005140 JAI47174.1 GDIQ01172818 JAK78907.1 GDRN01074321 JAI63263.1 GDIP01056408 JAM47307.1 CVRI01000001 CRK86494.1 GDIP01012441 JAM91274.1 GDIQ01189412 JAK62313.1 LJIG01022770 KRT78806.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000075903
UP000075882
UP000007062
+ More
UP000075840 UP000030765 UP000008820 UP000075900 UP000075886 UP000075920 UP000075885 UP000075880 UP000075881 UP000092462 UP000076408 UP000076407 UP000075902 UP000075884 UP000069272 UP000095300 UP000002320 UP000075883 UP000069940 UP000249989 UP000037069 UP000095301 UP000078200 UP000092444 UP000092445 UP000092460 UP000009192 UP000001070 UP000000673 UP000008792 UP000001819 UP000008744 UP000092553 UP000092461 UP000091820 UP000092443 UP000007798 UP000008711 UP000002282 UP000007801 UP000192221 UP000268350 UP000000803 UP000001292 UP000005203 UP000076502 UP000053825 UP000000311 UP000053105 UP000078542 UP000005205 UP000007755 UP000078540 UP000078541 UP000008237 UP000078492 UP000036403 UP000242457 UP000002358 UP000000305 UP000215335 UP000076858 UP000183832
UP000075840 UP000030765 UP000008820 UP000075900 UP000075886 UP000075920 UP000075885 UP000075880 UP000075881 UP000092462 UP000076408 UP000076407 UP000075902 UP000075884 UP000069272 UP000095300 UP000002320 UP000075883 UP000069940 UP000249989 UP000037069 UP000095301 UP000078200 UP000092444 UP000092445 UP000092460 UP000009192 UP000001070 UP000000673 UP000008792 UP000001819 UP000008744 UP000092553 UP000092461 UP000091820 UP000092443 UP000007798 UP000008711 UP000002282 UP000007801 UP000192221 UP000268350 UP000000803 UP000001292 UP000005203 UP000076502 UP000053825 UP000000311 UP000053105 UP000078542 UP000005205 UP000007755 UP000078540 UP000078541 UP000008237 UP000078492 UP000036403 UP000242457 UP000002358 UP000000305 UP000215335 UP000076858 UP000183832
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JD65
A0A194QFW7
A0A1S4FWV3
A0A336LH25
A0A212FIW4
A0A182URL5
+ More
A0A182L660 Q7Q1S5 A0A182ICS7 A0A084VWB7 Q16LU3 A0A182RGQ2 A0A182QU08 A0A182VTW5 A0A182PKV1 A0A182JFI1 A0A1Q3FRX6 A0A182JNR7 A0A1B0DIM0 A0A182Y8B4 A0A182WZE0 A0A182TN75 A0A182NAX9 A0A182F4D4 A0A1I8QB07 B0WCJ3 A0A182MCR0 A0A182GCT3 A0A0L0CI44 A0A1I8N9U6 A0A1A9UTW9 A0A1B0FMP5 A0A1A9Z5X8 A0A1B0BDS9 V5GWS5 A0A1Q3FRW5 B4KEU3 B4JDF0 W5J4J0 B4LU37 Q29LH5 B4GPY3 A0A0M4EEJ2 A0A1B0CDU0 A0A1A9WUC1 A0A1A9XYT0 B4MZJ1 B3N8C2 B4P311 B3MN94 A0A1W4VYB8 A0A3B0JC14 Q9VPX2 B4ID58 W8BWB5 A0A0J9QTR8 A0A0A1WZ82 A0A034WUJ8 T1DKQ2 A0A088A6U6 A0A154PL53 A0A0L7QLU6 A0A310SFK0 E2AHU1 A0A0N0U7D6 T1PBF5 A0A151IAL0 A0A158NEF3 F4WVK5 A0A195BV40 A0A1Y1NGT2 A0A195FL16 E2BIX9 A0A195EP60 A0A0J7KZ16 A0A0T6AZQ5 A0A2A3E9L3 K7IRU9 E9GHP0 A0A0P5ZVD2 A0A232EHZ9 A0A162D4Z8 A0A0P5QI90 A0A0P4ZW54 A0A0P5KF83 A0A0P6IKX3 A0A0P5G067 A0A0P5GR19 B4KEU4 E9GHN7 E9GKW4 E9GHN9 A0A0K8W7Z8 A0A0P5LAA3 A0A0P4WPA4 A0A0P5YKS6 A0A1J1HJ49 A0A0P6BRB5 A0A0P5KMP1 A0A0T6AUS9
A0A182L660 Q7Q1S5 A0A182ICS7 A0A084VWB7 Q16LU3 A0A182RGQ2 A0A182QU08 A0A182VTW5 A0A182PKV1 A0A182JFI1 A0A1Q3FRX6 A0A182JNR7 A0A1B0DIM0 A0A182Y8B4 A0A182WZE0 A0A182TN75 A0A182NAX9 A0A182F4D4 A0A1I8QB07 B0WCJ3 A0A182MCR0 A0A182GCT3 A0A0L0CI44 A0A1I8N9U6 A0A1A9UTW9 A0A1B0FMP5 A0A1A9Z5X8 A0A1B0BDS9 V5GWS5 A0A1Q3FRW5 B4KEU3 B4JDF0 W5J4J0 B4LU37 Q29LH5 B4GPY3 A0A0M4EEJ2 A0A1B0CDU0 A0A1A9WUC1 A0A1A9XYT0 B4MZJ1 B3N8C2 B4P311 B3MN94 A0A1W4VYB8 A0A3B0JC14 Q9VPX2 B4ID58 W8BWB5 A0A0J9QTR8 A0A0A1WZ82 A0A034WUJ8 T1DKQ2 A0A088A6U6 A0A154PL53 A0A0L7QLU6 A0A310SFK0 E2AHU1 A0A0N0U7D6 T1PBF5 A0A151IAL0 A0A158NEF3 F4WVK5 A0A195BV40 A0A1Y1NGT2 A0A195FL16 E2BIX9 A0A195EP60 A0A0J7KZ16 A0A0T6AZQ5 A0A2A3E9L3 K7IRU9 E9GHP0 A0A0P5ZVD2 A0A232EHZ9 A0A162D4Z8 A0A0P5QI90 A0A0P4ZW54 A0A0P5KF83 A0A0P6IKX3 A0A0P5G067 A0A0P5GR19 B4KEU4 E9GHN7 E9GKW4 E9GHN9 A0A0K8W7Z8 A0A0P5LAA3 A0A0P4WPA4 A0A0P5YKS6 A0A1J1HJ49 A0A0P6BRB5 A0A0P5KMP1 A0A0T6AUS9
PDB
6E9O
E-value=4.5873e-08,
Score=139
Ontologies
GO
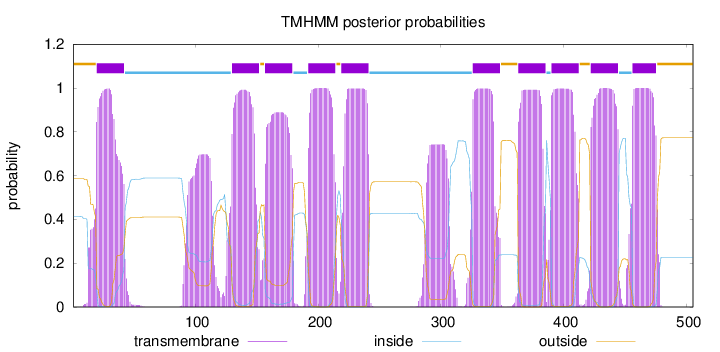
Topology
Length:
505
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
247.82694
Exp number, first 60 AAs:
22.711
Total prob of N-in:
0.41295
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 129
TMhelix
130 - 152
outside
153 - 156
TMhelix
157 - 179
inside
180 - 191
TMhelix
192 - 214
outside
215 - 218
TMhelix
219 - 241
inside
242 - 325
TMhelix
326 - 348
outside
349 - 362
TMhelix
363 - 385
inside
386 - 389
TMhelix
390 - 412
outside
413 - 421
TMhelix
422 - 444
inside
445 - 455
TMhelix
456 - 475
outside
476 - 505
Population Genetic Test Statistics
Pi
219.202619
Theta
172.316558
Tajima's D
0.795493
CLR
0.019193
CSRT
0.605719714014299
Interpretation
Uncertain
