Pre Gene Modal
BGIBMGA007475
Annotation
PREDICTED:_5-methylcytosine_rRNA_methyltransferase_NSUN4_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.2
Sequence
CDS
ATGCTTCAAATAACAAATTTGATCAAGAAAGGGCCTACAACTTGTGCTCACTTATTAAGACAATCTCGACATAAATCTAAAACTCATTGGGCAAAGTTAAGAAAAAAGACCGGCCCTAAATACAAGGCAATGAATCATTTTGATGAATTTTACGGGTCCGTTTTCGGTAAGAAATGGGATCCCATGAAACGAGCATTATTACAAGGCCCGAAGTATGTAGCCGTGATCAACAATTACGGAGACGCCGAAGAGACTATGGAGTATCTTACGAATAGAGGTGCACATTGCCTAAAGAAACTAATAACAGTCCAGGAAGATTTTCATAAACAATATTCTGTGAACACAACAACCTCTGATCAAGAAGCACCAGTATACATAAATAAATTACAGCAATTAGCAGAGAAGGCACATTCTGATGAAATATCCAGTATTTATCCGAATACTGATAATAAGCCTGAAAGACTCGACTTCAAAGACGATGATCAAATTTCTGAAAGAAAGGAAATTACGGAGCAGAAGGAACCAATAAACAAATCATTAGATCAGGCGATAGCAGATGCTGTCATAGATGAGTCTCGAATGATTGACCCATCTATAGGCTTGACCTCAGAAGCGTTGTATCAGTATTTACCAGCAACTAAACTTAAAGGCATGAATGACTGGGTACCAGAGTCTACACACTATTCTTATTATTCAAAAATAGTTGATTTCCCCTTAACTATCGAGCCCGAGACTGAATTGCAGTACCCGGAACACCTGAAAGTGTTCACTTATGAAATGGACAGTGAACTTAAAATGTTCCCCGAACCGAAGAGATCACAAACAGGCGTGTTTAACTACTACCCCATGGATTGTGGCAGCGTGTTAGCCGTGCTAGCCCTGGGGTTGAGAGCCGGCGACAGGGTCCTGGACTTGTGCTCTGCTCCCGGGGGCAAGGCGCTGGTCGCGCTGCAGACCTTGTTACCGGACGTGGTCATATGTAACGACGTGTCTATATCTAGATCGAACAGAACACAAAGGGTGTTCAGCGACTATCTGTTCGATTTCGAGACGGGCAACAAGTGGCGGGAACGCGTCATCTTCAAAAGGGTCGACGGCAGAATGTTCACAGACGACCACGGCTTCGATAAGGTTCTAGTGGACGTGCCGTGCACTACGGACCGACACTCTGTGACCGAGGATGACAACAACATCTTCAGGCCGGACCGGGTGAAGGAGAGGCTCAAGATACCGGAGCTCCAGTCGCACTTGTTGGCAAACGCGCTTCGTTTAACGAAGGTCGGGGGGACGGTCGTGTACTCCACGTGCTCGCTCAGCCCCATACAGAACGACGGGGTGGTGCACATGGCCCTCAAGCAAGTCTTCGAGAGCAACGACATCATCGCCACCGTCGTAGATTTAACTCCAGCGGTCGCCGGCCTCAGCAGCACCCTGACGCTGGGCGCGGGCGCTGCGGCCCCCAAGTACGGGCAGCTCGTGACTCCGTCGCTTTCCGCCAACTTCGGGCCGTCGTACATCGCGAAACTTTTACGAATTAAATAA
Protein
MLQITNLIKKGPTTCAHLLRQSRHKSKTHWAKLRKKTGPKYKAMNHFDEFYGSVFGKKWDPMKRALLQGPKYVAVINNYGDAEETMEYLTNRGAHCLKKLITVQEDFHKQYSVNTTTSDQEAPVYINKLQQLAEKAHSDEISSIYPNTDNKPERLDFKDDDQISERKEITEQKEPINKSLDQAIADAVIDESRMIDPSIGLTSEALYQYLPATKLKGMNDWVPESTHYSYYSKIVDFPLTIEPETELQYPEHLKVFTYEMDSELKMFPEPKRSQTGVFNYYPMDCGSVLAVLALGLRAGDRVLDLCSAPGGKALVALQTLLPDVVICNDVSISRSNRTQRVFSDYLFDFETGNKWRERVIFKRVDGRMFTDDHGFDKVLVDVPCTTDRHSVTEDDNNIFRPDRVKERLKIPELQSHLLANALRLTKVGGTVVYSTCSLSPIQNDGVVHMALKQVFESNDIIATVVDLTPAVAGLSSTLTLGAGAAAPKYGQLVTPSLSANFGPSYIAKLLRIK
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. RsmB/NOP family.
Uniprot
H9JD78
A0A212FIV6
A0A2W1BPB8
A0A194QBH5
A0A0L7LJ32
A0A067R711
+ More
B3MN96 A0A0L0CIA8 A0A2J7QGN6 B4P312 B4ID59 A0A1W4WA59 B3N8C3 A0A1A9UTX1 Q9VPX3 A0A1A9XYT4 A0A1B0FML8 A0A1A9Z5Y0 B4MZJ2 A0A0M3QTB2 A0A1B0BDS8 B4KEU5 A0A0J9QTM3 A0A034WSU4 A0A0K8TW15 A0A1I8P3U3 B4LU36 A0A1I8NEZ8 A0A182P4P6 B4JDF1 D6WUZ7 A0A1Y1L176 A0A182KEE1 A0A3B0K8Q7 A0A182L033 A0A182V6N2 A0A0C9QZD8 A0A2H8TEQ7 B0VZZ5 B4GPY5 Q29LH4 W8C9M7 A0A1S4GYI0 A0A182IBU5 A0A182X184 A0A0P4VRK4 A0A1Q3EYA4 A0A1B0CDT9 T1I219 A0A182GCT4 Q5TQ80 Q16LU4 A0A2S2PVU4 A0A1L8DSR0 A0A182RCA0 A0A182JHT5 A0A023F1F7 J9JKX0 A0A182U3X8 A0A0A9WF62 A0A1J1HES6 A0A182VTQ1 A0A0K8T721 A0A026WRL7 A0A146LY16 A0A0L7QLS4 W5JE37 A0A182NB43 A0A182Y8G6 A0A182QRU7 A0A182ME90 A0A182F2M2 A0A151XJF3 A0A084VBE6 A0A195FMB6 A0A151IAE6 A0A195ENE9 A0A1B6J4V4 A0A1B0DIL9 J3JTZ9 U4UGI3 A0A088A6Q5 B0XLM9 A0A195BWZ2 E2BIY0 A0A0R3NTX1 F4WVK4 A0A158NEF4 A0A1W4WRA5 N6UM02 K7IRU8 A0A2A3EAB5 A0A232EHZ2 E2AHU0 A0A1B6LM09 A0A154PJH7 A0A0V0GBX4 A0A0M9A9N1
B3MN96 A0A0L0CIA8 A0A2J7QGN6 B4P312 B4ID59 A0A1W4WA59 B3N8C3 A0A1A9UTX1 Q9VPX3 A0A1A9XYT4 A0A1B0FML8 A0A1A9Z5Y0 B4MZJ2 A0A0M3QTB2 A0A1B0BDS8 B4KEU5 A0A0J9QTM3 A0A034WSU4 A0A0K8TW15 A0A1I8P3U3 B4LU36 A0A1I8NEZ8 A0A182P4P6 B4JDF1 D6WUZ7 A0A1Y1L176 A0A182KEE1 A0A3B0K8Q7 A0A182L033 A0A182V6N2 A0A0C9QZD8 A0A2H8TEQ7 B0VZZ5 B4GPY5 Q29LH4 W8C9M7 A0A1S4GYI0 A0A182IBU5 A0A182X184 A0A0P4VRK4 A0A1Q3EYA4 A0A1B0CDT9 T1I219 A0A182GCT4 Q5TQ80 Q16LU4 A0A2S2PVU4 A0A1L8DSR0 A0A182RCA0 A0A182JHT5 A0A023F1F7 J9JKX0 A0A182U3X8 A0A0A9WF62 A0A1J1HES6 A0A182VTQ1 A0A0K8T721 A0A026WRL7 A0A146LY16 A0A0L7QLS4 W5JE37 A0A182NB43 A0A182Y8G6 A0A182QRU7 A0A182ME90 A0A182F2M2 A0A151XJF3 A0A084VBE6 A0A195FMB6 A0A151IAE6 A0A195ENE9 A0A1B6J4V4 A0A1B0DIL9 J3JTZ9 U4UGI3 A0A088A6Q5 B0XLM9 A0A195BWZ2 E2BIY0 A0A0R3NTX1 F4WVK4 A0A158NEF4 A0A1W4WRA5 N6UM02 K7IRU8 A0A2A3EAB5 A0A232EHZ2 E2AHU0 A0A1B6LM09 A0A154PJH7 A0A0V0GBX4 A0A0M9A9N1
Pubmed
19121390
22118469
28756777
26354079
26227816
24845553
+ More
17994087 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 25315136 18362917 19820115 28004739 20966253 15632085 24495485 12364791 27129103 26483478 17510324 25474469 25401762 24508170 30249741 26823975 20920257 23761445 25244985 24438588 22516182 23537049 20798317 21719571 21347285 20075255 28648823
17994087 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 25315136 18362917 19820115 28004739 20966253 15632085 24495485 12364791 27129103 26483478 17510324 25474469 25401762 24508170 30249741 26823975 20920257 23761445 25244985 24438588 22516182 23537049 20798317 21719571 21347285 20075255 28648823
EMBL
BABH01036245
AGBW02008325
OWR53659.1
KZ150100
PZC73523.1
KQ459249
+ More
KPJ02340.1 JTDY01000989 KOB75186.1 KK852699 KDR18203.1 CH902620 EDV31053.1 JRES01000438 KNC31229.1 NEVH01014361 PNF27738.1 CM000157 EDW87219.1 CH480829 EDW45485.1 CH954177 EDV57310.1 AE014134 AY058685 AAF51412.1 AAL13914.1 CCAG010018214 CH963920 EDW77777.1 CP012523 ALC38579.1 JXJN01012663 CH933807 EDW12995.1 CM002910 KMY87427.1 GAKP01002105 GAKP01002104 JAC56848.1 GDHF01033645 GDHF01017139 JAI18669.1 JAI35175.1 CH940649 EDW65089.1 CH916368 EDW03321.1 KQ971357 EFA08513.1 GEZM01073753 JAV64857.1 OUUW01000004 SPP79918.1 GBYB01000001 JAG69768.1 GFXV01000759 MBW12564.1 DS231816 EDS37567.1 CH479187 EDW39657.1 CH379060 EAL34071.1 GAMC01006506 JAC00050.1 AAAB01008964 APCN01000743 GDKW01001546 JAI55049.1 GFDL01014765 JAV20280.1 AJWK01008229 AJWK01008230 ACPB03000292 JXUM01054804 KQ561837 KXJ77388.1 EAL39544.3 CH477890 EAT35305.1 GGMS01000277 MBY69480.1 GFDF01004729 JAV09355.1 GBBI01003589 JAC15123.1 ABLF02040227 GBHO01038487 GBHO01038486 JAG05117.1 JAG05118.1 CVRI01000001 CRK86493.1 GBRD01004483 JAG61338.1 KK107119 QOIP01000009 EZA58657.1 RLU18276.1 GDHC01007208 GDHC01006111 JAQ11421.1 JAQ12518.1 KQ414905 KOC59598.1 ADMH02001492 ETN62326.1 AXCN02000644 AXCM01000666 KQ982080 KYQ60408.1 ATLV01006582 ATLV01006583 KE524368 KFB35290.1 KQ981490 KYN41109.1 KQ978221 KYM96279.1 KQ978625 KYN29676.1 GECU01013515 JAS94191.1 AJVK01006145 AJVK01006146 BT126710 AEE61672.1 KB632286 ERL91453.1 DS234577 EDS34468.1 KQ976401 KYM92488.1 GL448543 EFN84325.1 KRT04441.1 GL888387 EGI61825.1 ADTU01013276 APGK01018190 KB740071 ENN81691.1 KZ288312 PBC28414.1 NNAY01004388 OXU17965.1 GL439579 EFN67019.1 GEBQ01015202 JAT24775.1 KQ434936 KZC12019.1 GECL01000648 JAP05476.1 KQ435709 KOX80057.1
KPJ02340.1 JTDY01000989 KOB75186.1 KK852699 KDR18203.1 CH902620 EDV31053.1 JRES01000438 KNC31229.1 NEVH01014361 PNF27738.1 CM000157 EDW87219.1 CH480829 EDW45485.1 CH954177 EDV57310.1 AE014134 AY058685 AAF51412.1 AAL13914.1 CCAG010018214 CH963920 EDW77777.1 CP012523 ALC38579.1 JXJN01012663 CH933807 EDW12995.1 CM002910 KMY87427.1 GAKP01002105 GAKP01002104 JAC56848.1 GDHF01033645 GDHF01017139 JAI18669.1 JAI35175.1 CH940649 EDW65089.1 CH916368 EDW03321.1 KQ971357 EFA08513.1 GEZM01073753 JAV64857.1 OUUW01000004 SPP79918.1 GBYB01000001 JAG69768.1 GFXV01000759 MBW12564.1 DS231816 EDS37567.1 CH479187 EDW39657.1 CH379060 EAL34071.1 GAMC01006506 JAC00050.1 AAAB01008964 APCN01000743 GDKW01001546 JAI55049.1 GFDL01014765 JAV20280.1 AJWK01008229 AJWK01008230 ACPB03000292 JXUM01054804 KQ561837 KXJ77388.1 EAL39544.3 CH477890 EAT35305.1 GGMS01000277 MBY69480.1 GFDF01004729 JAV09355.1 GBBI01003589 JAC15123.1 ABLF02040227 GBHO01038487 GBHO01038486 JAG05117.1 JAG05118.1 CVRI01000001 CRK86493.1 GBRD01004483 JAG61338.1 KK107119 QOIP01000009 EZA58657.1 RLU18276.1 GDHC01007208 GDHC01006111 JAQ11421.1 JAQ12518.1 KQ414905 KOC59598.1 ADMH02001492 ETN62326.1 AXCN02000644 AXCM01000666 KQ982080 KYQ60408.1 ATLV01006582 ATLV01006583 KE524368 KFB35290.1 KQ981490 KYN41109.1 KQ978221 KYM96279.1 KQ978625 KYN29676.1 GECU01013515 JAS94191.1 AJVK01006145 AJVK01006146 BT126710 AEE61672.1 KB632286 ERL91453.1 DS234577 EDS34468.1 KQ976401 KYM92488.1 GL448543 EFN84325.1 KRT04441.1 GL888387 EGI61825.1 ADTU01013276 APGK01018190 KB740071 ENN81691.1 KZ288312 PBC28414.1 NNAY01004388 OXU17965.1 GL439579 EFN67019.1 GEBQ01015202 JAT24775.1 KQ434936 KZC12019.1 GECL01000648 JAP05476.1 KQ435709 KOX80057.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000037510
UP000027135
UP000007801
+ More
UP000037069 UP000235965 UP000002282 UP000001292 UP000192221 UP000008711 UP000078200 UP000000803 UP000092443 UP000092444 UP000092445 UP000007798 UP000092553 UP000092460 UP000009192 UP000095300 UP000008792 UP000095301 UP000075885 UP000001070 UP000007266 UP000075881 UP000268350 UP000075882 UP000075903 UP000002320 UP000008744 UP000001819 UP000075840 UP000076407 UP000092461 UP000015103 UP000069940 UP000249989 UP000007062 UP000008820 UP000075900 UP000075880 UP000007819 UP000075902 UP000183832 UP000075920 UP000053097 UP000279307 UP000053825 UP000000673 UP000075884 UP000076408 UP000075886 UP000075883 UP000069272 UP000075809 UP000030765 UP000078541 UP000078542 UP000078492 UP000092462 UP000030742 UP000005203 UP000078540 UP000008237 UP000007755 UP000005205 UP000192223 UP000019118 UP000002358 UP000242457 UP000215335 UP000000311 UP000076502 UP000053105
UP000037069 UP000235965 UP000002282 UP000001292 UP000192221 UP000008711 UP000078200 UP000000803 UP000092443 UP000092444 UP000092445 UP000007798 UP000092553 UP000092460 UP000009192 UP000095300 UP000008792 UP000095301 UP000075885 UP000001070 UP000007266 UP000075881 UP000268350 UP000075882 UP000075903 UP000002320 UP000008744 UP000001819 UP000075840 UP000076407 UP000092461 UP000015103 UP000069940 UP000249989 UP000007062 UP000008820 UP000075900 UP000075880 UP000007819 UP000075902 UP000183832 UP000075920 UP000053097 UP000279307 UP000053825 UP000000673 UP000075884 UP000076408 UP000075886 UP000075883 UP000069272 UP000075809 UP000030765 UP000078541 UP000078542 UP000078492 UP000092462 UP000030742 UP000005203 UP000078540 UP000008237 UP000007755 UP000005205 UP000192223 UP000019118 UP000002358 UP000242457 UP000215335 UP000000311 UP000076502 UP000053105
Pfam
PF01189 Methyltr_RsmB-F
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9JD78
A0A212FIV6
A0A2W1BPB8
A0A194QBH5
A0A0L7LJ32
A0A067R711
+ More
B3MN96 A0A0L0CIA8 A0A2J7QGN6 B4P312 B4ID59 A0A1W4WA59 B3N8C3 A0A1A9UTX1 Q9VPX3 A0A1A9XYT4 A0A1B0FML8 A0A1A9Z5Y0 B4MZJ2 A0A0M3QTB2 A0A1B0BDS8 B4KEU5 A0A0J9QTM3 A0A034WSU4 A0A0K8TW15 A0A1I8P3U3 B4LU36 A0A1I8NEZ8 A0A182P4P6 B4JDF1 D6WUZ7 A0A1Y1L176 A0A182KEE1 A0A3B0K8Q7 A0A182L033 A0A182V6N2 A0A0C9QZD8 A0A2H8TEQ7 B0VZZ5 B4GPY5 Q29LH4 W8C9M7 A0A1S4GYI0 A0A182IBU5 A0A182X184 A0A0P4VRK4 A0A1Q3EYA4 A0A1B0CDT9 T1I219 A0A182GCT4 Q5TQ80 Q16LU4 A0A2S2PVU4 A0A1L8DSR0 A0A182RCA0 A0A182JHT5 A0A023F1F7 J9JKX0 A0A182U3X8 A0A0A9WF62 A0A1J1HES6 A0A182VTQ1 A0A0K8T721 A0A026WRL7 A0A146LY16 A0A0L7QLS4 W5JE37 A0A182NB43 A0A182Y8G6 A0A182QRU7 A0A182ME90 A0A182F2M2 A0A151XJF3 A0A084VBE6 A0A195FMB6 A0A151IAE6 A0A195ENE9 A0A1B6J4V4 A0A1B0DIL9 J3JTZ9 U4UGI3 A0A088A6Q5 B0XLM9 A0A195BWZ2 E2BIY0 A0A0R3NTX1 F4WVK4 A0A158NEF4 A0A1W4WRA5 N6UM02 K7IRU8 A0A2A3EAB5 A0A232EHZ2 E2AHU0 A0A1B6LM09 A0A154PJH7 A0A0V0GBX4 A0A0M9A9N1
B3MN96 A0A0L0CIA8 A0A2J7QGN6 B4P312 B4ID59 A0A1W4WA59 B3N8C3 A0A1A9UTX1 Q9VPX3 A0A1A9XYT4 A0A1B0FML8 A0A1A9Z5Y0 B4MZJ2 A0A0M3QTB2 A0A1B0BDS8 B4KEU5 A0A0J9QTM3 A0A034WSU4 A0A0K8TW15 A0A1I8P3U3 B4LU36 A0A1I8NEZ8 A0A182P4P6 B4JDF1 D6WUZ7 A0A1Y1L176 A0A182KEE1 A0A3B0K8Q7 A0A182L033 A0A182V6N2 A0A0C9QZD8 A0A2H8TEQ7 B0VZZ5 B4GPY5 Q29LH4 W8C9M7 A0A1S4GYI0 A0A182IBU5 A0A182X184 A0A0P4VRK4 A0A1Q3EYA4 A0A1B0CDT9 T1I219 A0A182GCT4 Q5TQ80 Q16LU4 A0A2S2PVU4 A0A1L8DSR0 A0A182RCA0 A0A182JHT5 A0A023F1F7 J9JKX0 A0A182U3X8 A0A0A9WF62 A0A1J1HES6 A0A182VTQ1 A0A0K8T721 A0A026WRL7 A0A146LY16 A0A0L7QLS4 W5JE37 A0A182NB43 A0A182Y8G6 A0A182QRU7 A0A182ME90 A0A182F2M2 A0A151XJF3 A0A084VBE6 A0A195FMB6 A0A151IAE6 A0A195ENE9 A0A1B6J4V4 A0A1B0DIL9 J3JTZ9 U4UGI3 A0A088A6Q5 B0XLM9 A0A195BWZ2 E2BIY0 A0A0R3NTX1 F4WVK4 A0A158NEF4 A0A1W4WRA5 N6UM02 K7IRU8 A0A2A3EAB5 A0A232EHZ2 E2AHU0 A0A1B6LM09 A0A154PJH7 A0A0V0GBX4 A0A0M9A9N1
PDB
4FP9
E-value=4.67249e-43,
Score=440
Ontologies
GO
Topology
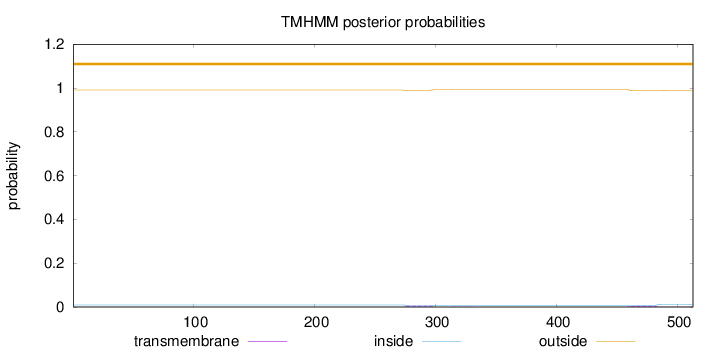
Length:
513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28618
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00932
outside
1 - 513
Population Genetic Test Statistics
Pi
233.292259
Theta
169.235848
Tajima's D
1.526833
CLR
0.019096
CSRT
0.795560221988901
Interpretation
Uncertain
