Gene
KWMTBOMO01139 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007473
Annotation
PREDICTED:_tudor_and_KH_domain-containing_protein_[Bombyx_mori]
Full name
Tudor and KH domain-containing protein homolog
+ More
Katanin p80 WD40 repeat-containing subunit B1
Katanin p80 WD40 repeat-containing subunit B1
Alternative Name
Partner of PIWIs protein
p80 katanin
p80 katanin
Location in the cell
Nuclear Reliability : 2.511
Sequence
CDS
ATGTCATTGAACACAAAATTAGCTTTGCCCATTGCTTTGGGCTTGTCCCTCGTGACTGTAACAGCATTTGTGGCCTACTATGTCCTCAAGAGAGATGAAGAAGAAAACGATAACAAGACAGTCAAGATTACTAAAATAAATACAATTGAAATTCATGTTCCTAAAAGCATTGTTCCAGCTCTGATTGGTCGCAATGGCTCCAACATTAAGGACCTTCAGAAGAAATCTGGTGCACAGATCCATTTCAAAAAGTTCACCGACCAGGACTATGATGTGTGTGTAGTCAGAGGACGAGCTGACACTACTCAGCTGGCTGAAACTTTAATACATGACTTTATCAAGCAACAGCCAACAATTATGTCAGAAAGTATTACTGTGCCAAGCTGGTCTTGTGGGAGAATAATTGGATCTGGAGGTGAGAACGTCAATGATATTAGCCATCGCAGTGGCGCTCGCGTCAAGGTCGAAAGTCCTAAAAGTACAGATAAAGTTGCTGAGCATCTAGTGACATTCCGTGGTACCAAGGAACAGATTGAAGTCGCCAAGAAACTTGTTGAGAACTGCATATCTTTAGAGCGGTGTAGACGGGAGATAGAGCAGTCGAAGCGTCCGCCCCGGCACAGCTCGTCGCCCCCCTCCCCCTGCCCCTCGCCCGGCGACCGCGACGCGGACGCGGACGCACAAGAGACAACGGGACCCAGTATCGAAGTGTACGTGTCGGCGGTTTCATCTCCTTCCCGTTTCTGGGTGCAGTTCGTAGGGCCCCAGGTCGCTCAGCTTGACGACCTAGTCGCTCACATGACCGAGTATTACTCTAAGAAGGAGAACAGAGAAGCGCATACCTTACGCCACGTGTCCGTTGGGCAAGTGGTGGCGGCGGTGTTCCGACACGACGGCCGCTGGTACCGGGCAAGGGTGCACGACATCAGGCCTAACGAGTTCGATTCATCGCAACAGGTAGCCGACGTGTTCTATCTGGACTACGGCGACAGCGAGTACGTGGCTACGCACGAGCTGTGCGAGCTAAGAGCGGACCTATTAAGGCTGCGGTTCCAGGCCATGGAGTGCTTCCTGGCTGGCGTCCGGCCCGCCAGCGGCGAGGAGGCCGTGTCTCCGTCTGGCCAGCGTTGGGACAAGTGGCATCCACAGGCCGTCGAGAGGTTCGAAGAACTTACCCAGGTAGCGAGGTGGAAGGCCCTGGTCTCCCGGACCTGCACGTACAAGAAGACGGCGACCGCTGAAGGAGAGAAGGACAAAGAGATACCTGGGATCAAACTGTTCGATGTTACTGATGAGGGCGAGCTGGACGTGGGCGCGGTGCTGGTGGCGGAGGGGTGGGCGGTGGCGGGCCCCGCCCCCTCCCCGCGCCCCTCGCCCCCGCGCGGACACACCACGCCCTTCGGAGACCTCAGCAAGTCCAAAGTACTCAGTATGACCGGCGGTGGTGGACGATCATCGTCCGTTCCAAAGGATCACAAAGACGATGGCAGCAATGTGCTCACCGTTGAAGGCGATGACTCCAAAGACAAAGACGGAATAACAGCATCGAAATCACTAGCCTCCGGTCTTGAGAAGTCTGACCGCCATCCGTTATCGATATCGAACTTCGATCTATCGTATCCCGACCCTTCGAGGAACAAACAACTGAACGGCTCCGATGATTTCTTGCACGGCGAGCGGCAAAATATCGATAACGAAATCACTATCGATACATTAGTTCCGCCGTCTCCCCTCTCTAACGCTCCGATACTGTCGAAAACGCTTCAAGACGAATTCAAGGCCAACATGAACCGAATAGATTCGCACCACAGTAATCTCGAAAACCTCGGTAAGTCCGCTTTTGAAAAGTGA
Protein
MSLNTKLALPIALGLSLVTVTAFVAYYVLKRDEEENDNKTVKITKINTIEIHVPKSIVPALIGRNGSNIKDLQKKSGAQIHFKKFTDQDYDVCVVRGRADTTQLAETLIHDFIKQQPTIMSESITVPSWSCGRIIGSGGENVNDISHRSGARVKVESPKSTDKVAEHLVTFRGTKEQIEVAKKLVENCISLERCRREIEQSKRPPRHSSSPPSPCPSPGDRDADADAQETTGPSIEVYVSAVSSPSRFWVQFVGPQVAQLDDLVAHMTEYYSKKENREAHTLRHVSVGQVVAAVFRHDGRWYRARVHDIRPNEFDSSQQVADVFYLDYGDSEYVATHELCELRADLLRLRFQAMECFLAGVRPASGEEAVSPSGQRWDKWHPQAVERFEELTQVARWKALVSRTCTYKKTATAEGEKDKEIPGIKLFDVTDEGELDVGAVLVAEGWAVAGPAPSPRPSPPRGHTTPFGDLSKSKVLSMTGGGGRSSSVPKDHKDDGSNVLTVEGDDSKDKDGITASKSLASGLEKSDRHPLSISNFDLSYPDPSRNKQLNGSDDFLHGERQNIDNEITIDTLVPPSPLSNAPILSKTLQDEFKANMNRIDSHHSNLENLGKSAFEK
Summary
Description
Participates in the primary piRNA biogenesis pathway and is required during spermatogenesis to repress transposable elements and prevent their mobilization, which is essential for the germline integrity (PubMed:23970546, PubMed:26919431). The piRNA metabolic process mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins (Siwi or Ago3) and govern the methylation and subsequent repression of transposons (PubMed:23970546, PubMed:26919431). Required for the final steps of primary piRNA biogenesis by participating in the processing of 31-37 nt intermediates into mature piRNAs: acts by recruiting the exonuclease PNLDC1/trimmer to Siwi-bound pre-piRNAs (PubMed:26919431).
Participates in a complex which severs microtubules in an ATP-dependent manner. May act to target the enzymatic subunit of this complex to sites of action such as the centrosome. Microtubule severing may promote rapid reorganization of cellular microtubule arrays and the release of microtubules from the centrosome following nucleation.
Involved in the piwi-interacting RNA (piRNA) metabolic process, which mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins, and governs the methylation and subsequent repression of transposons which is essential for germline integrity (PubMed:21447556, PubMed:29531043). Likely to act by recruiting Piwi proteins such as AGO3 and piwi to the piRNA biogenesis machinery in the nuage (PubMed:21447556, PubMed:29531043). Required for the final steps of primary piRNA biogenesis by participating in the 3' end-trimming of piwi-bound intermediates into mature piRNAs (PubMed:29531043).
Participates in a complex which severs microtubules in an ATP-dependent manner. May act to target the enzymatic subunit of this complex to sites of action such as the centrosome. Microtubule severing may promote rapid reorganization of cellular microtubule arrays and the release of microtubules from the centrosome following nucleation.
Involved in the piwi-interacting RNA (piRNA) metabolic process, which mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins, and governs the methylation and subsequent repression of transposons which is essential for germline integrity (PubMed:21447556, PubMed:29531043). Likely to act by recruiting Piwi proteins such as AGO3 and piwi to the piRNA biogenesis machinery in the nuage (PubMed:21447556, PubMed:29531043). Required for the final steps of primary piRNA biogenesis by participating in the 3' end-trimming of piwi-bound intermediates into mature piRNAs (PubMed:29531043).
Subunit
Interacts with (symmetrically methylated) Siwi (PubMed:23970546, PubMed:26919431). Interacts with (symmetrically methylated) Ago3 (PubMed:23970546). Interacts with PNLDC1/trimmer; interaction takes place on the mitochondrial surface and recruits PNLDC1/trimmer to Siwi-bound pre-piRNAs (PubMed:26919431).
Interacts with KATNA1. This interaction enhances the microtubule binding and severing activity of KATNA1 and also targets this activity to the centrosome.
Interacts (via C-terminus) with AGO3 (via the N-terminal region when symmetrically methylated on arginine residues); this interaction is RNA-independent and may be required for AGO3 localization to the nuage (PubMed:21447556). Interacts (via Tudor domain) with piwi (via N-terminus) (PubMed:21447556, PubMed:29531043). Interacts with tral and me31B (PubMed:21447556).
Interacts with KATNA1. This interaction enhances the microtubule binding and severing activity of KATNA1 and also targets this activity to the centrosome.
Interacts (via C-terminus) with AGO3 (via the N-terminal region when symmetrically methylated on arginine residues); this interaction is RNA-independent and may be required for AGO3 localization to the nuage (PubMed:21447556). Interacts (via Tudor domain) with piwi (via N-terminus) (PubMed:21447556, PubMed:29531043). Interacts with tral and me31B (PubMed:21447556).
Similarity
Belongs to the Tdrkh family.
Belongs to the WD repeat KATNB1 family.
Belongs to the XK family.
Belongs to the universal ribosomal protein uS3 family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Belongs to the WD repeat KATNB1 family.
Belongs to the XK family.
Belongs to the universal ribosomal protein uS3 family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Keywords
3D-structure
Complete proteome
Differentiation
Membrane
Mitochondrion
Mitochondrion outer membrane
Reference proteome
Repeat
RNA-binding
RNA-mediated gene silencing
Spermatogenesis
Transmembrane
Transmembrane helix
Cytoplasm
Nucleus
Feature
chain Tudor and KH domain-containing protein homolog
Uniprot
H9JD76
A0A1U6USP0
A0A194QA36
A0A0N1I9W3
A0A212FFX2
A0A2J7QI31
+ More
A0A0K8TRZ3 A0A1B0D8U1 J3JXU7 A0A1B6LIZ4 A0A067QZR2 A0A2P8Y4M7 A0A1I8P512 A0A023EU64 Q16K97 A0A1B0A7M9 A0A182GEX1 A0A0K8VNK3 A0A1Y1K1L9 A0A034VWN9 W8B6G7 A0A1Q3FJE6 D3TQM8 B0X0X8 A0A1B0GB97 A0A1B6FKZ6 A0A1B6G1S2 A0A1B6D308 A0A0T6AVE4 A0A1B0CI38 N6UGD8 A0A1A9YIS2 A0A182JJZ3 A0A1B0C1N4 B4KFL0 A0A1A9UNR6 E0VNB8 Q29MJ4 Q9VQ91 A0A1B6ITE1 B3MJA6 A0A1W4UTZ4 B4NW68 B4I2G5 B4G954 A0A0J9TDQ5 A0A182MGW3 A0A182JWC0 B4Q802 U5ENN8 B3N9H5 A0A088AD82 A0A182N8N1 A0A182PXE7 A0A182QH34 A0A1L8DLS9 A0A1J1IKM6 A0A1J1IKG4 A0A1L8DLF5 A0A182I0L2 Q7Q0T6 A0A224XPC3 A0A182XB16 A0A182UIK4 A0A1W4WRY2 A0A182UTS0 A0A182R7S0 A0A182YIK6 A0A182S883 A0A0C9PV82 A0A1S3KBL4 A0A084VQ04 V4A8S8
A0A0K8TRZ3 A0A1B0D8U1 J3JXU7 A0A1B6LIZ4 A0A067QZR2 A0A2P8Y4M7 A0A1I8P512 A0A023EU64 Q16K97 A0A1B0A7M9 A0A182GEX1 A0A0K8VNK3 A0A1Y1K1L9 A0A034VWN9 W8B6G7 A0A1Q3FJE6 D3TQM8 B0X0X8 A0A1B0GB97 A0A1B6FKZ6 A0A1B6G1S2 A0A1B6D308 A0A0T6AVE4 A0A1B0CI38 N6UGD8 A0A1A9YIS2 A0A182JJZ3 A0A1B0C1N4 B4KFL0 A0A1A9UNR6 E0VNB8 Q29MJ4 Q9VQ91 A0A1B6ITE1 B3MJA6 A0A1W4UTZ4 B4NW68 B4I2G5 B4G954 A0A0J9TDQ5 A0A182MGW3 A0A182JWC0 B4Q802 U5ENN8 B3N9H5 A0A088AD82 A0A182N8N1 A0A182PXE7 A0A182QH34 A0A1L8DLS9 A0A1J1IKM6 A0A1J1IKG4 A0A1L8DLF5 A0A182I0L2 Q7Q0T6 A0A224XPC3 A0A182XB16 A0A182UIK4 A0A1W4WRY2 A0A182UTS0 A0A182R7S0 A0A182YIK6 A0A182S883 A0A0C9PV82 A0A1S3KBL4 A0A084VQ04 V4A8S8
Pubmed
19121390
23970546
26919431
26354079
22118469
26369729
+ More
22516182 24845553 29403074 24945155 17510324 26483478 28004739 25348373 24495485 20353571 23537049 17994087 18057021 20566863 15632085 23185243 10731132 12537572 12537569 21447556 29531043 17550304 22936249 12364791 14747013 17210077 25244985 24438588 23254933
22516182 24845553 29403074 24945155 17510324 26483478 28004739 25348373 24495485 20353571 23537049 17994087 18057021 20566863 15632085 23185243 10731132 12537572 12537569 21447556 29531043 17550304 22936249 12364791 14747013 17210077 25244985 24438588 23254933
EMBL
BABH01036235
BABH01036236
BABH01036237
BABH01036238
KQ459249
KPJ02337.1
+ More
KQ460468 KPJ14587.1 AGBW02008752 OWR52635.1 NEVH01013964 PNF28213.1 GDAI01000680 JAI16923.1 AJVK01027734 BT128069 AEE63030.1 GEBQ01016332 JAT23645.1 KK852854 KDR15021.1 PYGN01000938 PSN39206.1 GAPW01000658 JAC12940.1 CH477971 EAT34718.1 JXUM01010565 KQ560311 KXJ83116.1 GDHF01030576 GDHF01024009 GDHF01019077 GDHF01011850 GDHF01001984 GDHF01001474 JAI21738.1 JAI28305.1 JAI33237.1 JAI40464.1 JAI50330.1 JAI50840.1 GEZM01095796 JAV55343.1 GAKP01012430 GAKP01012428 GAKP01012427 GAKP01012426 JAC46522.1 GAMC01012363 JAB94192.1 GFDL01007401 JAV27644.1 EZ423730 ADD20006.1 DS232248 EDS38370.1 CCAG010011564 GECZ01018918 GECZ01011322 JAS50851.1 JAS58447.1 GECZ01019313 GECZ01013395 GECZ01005951 JAS50456.1 JAS56374.1 JAS63818.1 GEDC01017234 JAS20064.1 LJIG01022720 KRT79064.1 AJWK01013029 AJWK01013030 APGK01036313 KB740939 KB631813 ENN77717.1 ERL86377.1 JXJN01024138 CH933807 EDW13125.1 KRG03586.1 KRG03587.1 DS235335 EEB14875.1 CH379060 EAL33700.1 KRT04125.1 AE014134 AY058408 GECU01028454 GECU01027109 GECU01017520 JAS79252.1 JAS80597.1 JAS90186.1 CH902620 EDV31316.1 KPU73314.1 KPU73315.1 CM000157 EDW87348.1 KRJ97127.1 KRJ97128.1 CH480820 EDW53960.1 CH479180 EDW28884.1 CM002910 KMY87613.1 KMY87614.1 KMY87615.1 KMY87616.1 AXCM01004445 CM000361 EDX03424.1 GANO01000519 JAB59352.1 CH954177 EDV57432.1 KQS69990.1 AXCN02001856 GFDF01006784 JAV07300.1 CVRI01000054 CRL00728.1 CRL00727.1 GFDF01006785 JAV07299.1 APCN01005219 AAAB01008980 EAA13859.4 GFTR01006413 JAW10013.1 GBYB01005303 JAG75070.1 ATLV01015102 KE525002 KFB40048.1 KB202544 ESO89711.1
KQ460468 KPJ14587.1 AGBW02008752 OWR52635.1 NEVH01013964 PNF28213.1 GDAI01000680 JAI16923.1 AJVK01027734 BT128069 AEE63030.1 GEBQ01016332 JAT23645.1 KK852854 KDR15021.1 PYGN01000938 PSN39206.1 GAPW01000658 JAC12940.1 CH477971 EAT34718.1 JXUM01010565 KQ560311 KXJ83116.1 GDHF01030576 GDHF01024009 GDHF01019077 GDHF01011850 GDHF01001984 GDHF01001474 JAI21738.1 JAI28305.1 JAI33237.1 JAI40464.1 JAI50330.1 JAI50840.1 GEZM01095796 JAV55343.1 GAKP01012430 GAKP01012428 GAKP01012427 GAKP01012426 JAC46522.1 GAMC01012363 JAB94192.1 GFDL01007401 JAV27644.1 EZ423730 ADD20006.1 DS232248 EDS38370.1 CCAG010011564 GECZ01018918 GECZ01011322 JAS50851.1 JAS58447.1 GECZ01019313 GECZ01013395 GECZ01005951 JAS50456.1 JAS56374.1 JAS63818.1 GEDC01017234 JAS20064.1 LJIG01022720 KRT79064.1 AJWK01013029 AJWK01013030 APGK01036313 KB740939 KB631813 ENN77717.1 ERL86377.1 JXJN01024138 CH933807 EDW13125.1 KRG03586.1 KRG03587.1 DS235335 EEB14875.1 CH379060 EAL33700.1 KRT04125.1 AE014134 AY058408 GECU01028454 GECU01027109 GECU01017520 JAS79252.1 JAS80597.1 JAS90186.1 CH902620 EDV31316.1 KPU73314.1 KPU73315.1 CM000157 EDW87348.1 KRJ97127.1 KRJ97128.1 CH480820 EDW53960.1 CH479180 EDW28884.1 CM002910 KMY87613.1 KMY87614.1 KMY87615.1 KMY87616.1 AXCM01004445 CM000361 EDX03424.1 GANO01000519 JAB59352.1 CH954177 EDV57432.1 KQS69990.1 AXCN02001856 GFDF01006784 JAV07300.1 CVRI01000054 CRL00728.1 CRL00727.1 GFDF01006785 JAV07299.1 APCN01005219 AAAB01008980 EAA13859.4 GFTR01006413 JAW10013.1 GBYB01005303 JAG75070.1 ATLV01015102 KE525002 KFB40048.1 KB202544 ESO89711.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000235965
UP000092462
+ More
UP000027135 UP000245037 UP000095300 UP000008820 UP000092445 UP000069940 UP000249989 UP000002320 UP000092444 UP000092461 UP000019118 UP000030742 UP000092443 UP000075880 UP000092460 UP000009192 UP000078200 UP000009046 UP000001819 UP000000803 UP000007801 UP000192221 UP000002282 UP000001292 UP000008744 UP000075883 UP000075881 UP000000304 UP000008711 UP000005203 UP000075884 UP000075885 UP000075886 UP000183832 UP000075840 UP000007062 UP000076407 UP000075902 UP000192223 UP000075903 UP000075900 UP000076408 UP000075901 UP000085678 UP000030765 UP000030746
UP000027135 UP000245037 UP000095300 UP000008820 UP000092445 UP000069940 UP000249989 UP000002320 UP000092444 UP000092461 UP000019118 UP000030742 UP000092443 UP000075880 UP000092460 UP000009192 UP000078200 UP000009046 UP000001819 UP000000803 UP000007801 UP000192221 UP000002282 UP000001292 UP000008744 UP000075883 UP000075881 UP000000304 UP000008711 UP000005203 UP000075884 UP000075885 UP000075886 UP000183832 UP000075840 UP000007062 UP000076407 UP000075902 UP000192223 UP000075903 UP000075900 UP000076408 UP000075901 UP000085678 UP000030765 UP000030746
Pfam
Interpro
IPR004088
KH_dom_type_1
+ More
IPR035437 SNase_OB-fold_sf
IPR036612 KH_dom_type_1_sf
IPR002999 Tudor
IPR004087 KH_dom
IPR001680 WD40_repeat
IPR024977 Apc4_WD40_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR028021 Katanin_C-terminal
IPR018629 XK-rel
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR026962 Katanin_bsu1
IPR005703 Ribosomal_S3_euk/arc
IPR036419 Ribosomal_S3_C_sf
IPR001351 Ribosomal_S3_C
IPR004044 KH_dom_type_2
IPR015946 KH_dom-like_a/b
IPR018280 Ribosomal_S3_CS
IPR009019 KH_sf_prok-type
IPR024518 DUF3421
IPR006616 DM9_repeat
IPR023395 Mt_carrier_dom_sf
IPR018108 Mitochondrial_sb/sol_carrier
IPR035437 SNase_OB-fold_sf
IPR036612 KH_dom_type_1_sf
IPR002999 Tudor
IPR004087 KH_dom
IPR001680 WD40_repeat
IPR024977 Apc4_WD40_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR028021 Katanin_C-terminal
IPR018629 XK-rel
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR026962 Katanin_bsu1
IPR005703 Ribosomal_S3_euk/arc
IPR036419 Ribosomal_S3_C_sf
IPR001351 Ribosomal_S3_C
IPR004044 KH_dom_type_2
IPR015946 KH_dom-like_a/b
IPR018280 Ribosomal_S3_CS
IPR009019 KH_sf_prok-type
IPR024518 DUF3421
IPR006616 DM9_repeat
IPR023395 Mt_carrier_dom_sf
IPR018108 Mitochondrial_sb/sol_carrier
SUPFAM
ProteinModelPortal
H9JD76
A0A1U6USP0
A0A194QA36
A0A0N1I9W3
A0A212FFX2
A0A2J7QI31
+ More
A0A0K8TRZ3 A0A1B0D8U1 J3JXU7 A0A1B6LIZ4 A0A067QZR2 A0A2P8Y4M7 A0A1I8P512 A0A023EU64 Q16K97 A0A1B0A7M9 A0A182GEX1 A0A0K8VNK3 A0A1Y1K1L9 A0A034VWN9 W8B6G7 A0A1Q3FJE6 D3TQM8 B0X0X8 A0A1B0GB97 A0A1B6FKZ6 A0A1B6G1S2 A0A1B6D308 A0A0T6AVE4 A0A1B0CI38 N6UGD8 A0A1A9YIS2 A0A182JJZ3 A0A1B0C1N4 B4KFL0 A0A1A9UNR6 E0VNB8 Q29MJ4 Q9VQ91 A0A1B6ITE1 B3MJA6 A0A1W4UTZ4 B4NW68 B4I2G5 B4G954 A0A0J9TDQ5 A0A182MGW3 A0A182JWC0 B4Q802 U5ENN8 B3N9H5 A0A088AD82 A0A182N8N1 A0A182PXE7 A0A182QH34 A0A1L8DLS9 A0A1J1IKM6 A0A1J1IKG4 A0A1L8DLF5 A0A182I0L2 Q7Q0T6 A0A224XPC3 A0A182XB16 A0A182UIK4 A0A1W4WRY2 A0A182UTS0 A0A182R7S0 A0A182YIK6 A0A182S883 A0A0C9PV82 A0A1S3KBL4 A0A084VQ04 V4A8S8
A0A0K8TRZ3 A0A1B0D8U1 J3JXU7 A0A1B6LIZ4 A0A067QZR2 A0A2P8Y4M7 A0A1I8P512 A0A023EU64 Q16K97 A0A1B0A7M9 A0A182GEX1 A0A0K8VNK3 A0A1Y1K1L9 A0A034VWN9 W8B6G7 A0A1Q3FJE6 D3TQM8 B0X0X8 A0A1B0GB97 A0A1B6FKZ6 A0A1B6G1S2 A0A1B6D308 A0A0T6AVE4 A0A1B0CI38 N6UGD8 A0A1A9YIS2 A0A182JJZ3 A0A1B0C1N4 B4KFL0 A0A1A9UNR6 E0VNB8 Q29MJ4 Q9VQ91 A0A1B6ITE1 B3MJA6 A0A1W4UTZ4 B4NW68 B4I2G5 B4G954 A0A0J9TDQ5 A0A182MGW3 A0A182JWC0 B4Q802 U5ENN8 B3N9H5 A0A088AD82 A0A182N8N1 A0A182PXE7 A0A182QH34 A0A1L8DLS9 A0A1J1IKM6 A0A1J1IKG4 A0A1L8DLF5 A0A182I0L2 Q7Q0T6 A0A224XPC3 A0A182XB16 A0A182UIK4 A0A1W4WRY2 A0A182UTS0 A0A182R7S0 A0A182YIK6 A0A182S883 A0A0C9PV82 A0A1S3KBL4 A0A084VQ04 V4A8S8
PDB
5VY1
E-value=5.09747e-127,
Score=1165
Ontologies
KEGG
GO
GO:0003723
GO:0030154
GO:0007283
GO:0005741
GO:0016021
GO:0031047
GO:1990511
GO:0016301
GO:0005874
GO:0008017
GO:0005813
GO:0000922
GO:0007049
GO:0051301
GO:0051013
GO:0008352
GO:0015935
GO:0006412
GO:0003735
GO:0070895
GO:0047485
GO:1903829
GO:0043186
GO:0005634
GO:0032991
GO:0000932
GO:0022857
GO:0003676
GO:0006303
GO:0004003
GO:0045901
GO:0045905
GO:0006334
GO:0003779
Topology
Subcellular location
Mitochondrion outer membrane
Localizes to the outer surface of mitochondria. With evidence from 5 publications.
Cytoplasm Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Cytoskeleton Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Microtubule organizing center Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Centrosome Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Spindle pole Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Spindle Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Membrane Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Nucleus Component of the meiotic nuage, also named P granule, a germ-cell-specific organelle required to repress transposon activity during meiosis. Expressed in the cytoplasm of germline stem cells, cyst cells, nurse cells and oocytes, with strong accumulation in region IIb germline cysts in the germarium. In post-germarium egg chambers accumulates in the perinuclear loci that appear to be the nuage. With evidence from 7 publications.
P-body Component of the meiotic nuage, also named P granule, a germ-cell-specific organelle required to repress transposon activity during meiosis. Expressed in the cytoplasm of germline stem cells, cyst cells, nurse cells and oocytes, with strong accumulation in region IIb germline cysts in the germarium. In post-germarium egg chambers accumulates in the perinuclear loci that appear to be the nuage. With evidence from 7 publications.
Cytoplasm Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Cytoskeleton Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Microtubule organizing center Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Centrosome Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Spindle pole Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Spindle Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Membrane Predominantly cytoplasmic. Localized to the interphase centrosome and mitotic spindle poles. With evidence from 1 publications.
Nucleus Component of the meiotic nuage, also named P granule, a germ-cell-specific organelle required to repress transposon activity during meiosis. Expressed in the cytoplasm of germline stem cells, cyst cells, nurse cells and oocytes, with strong accumulation in region IIb germline cysts in the germarium. In post-germarium egg chambers accumulates in the perinuclear loci that appear to be the nuage. With evidence from 7 publications.
P-body Component of the meiotic nuage, also named P granule, a germ-cell-specific organelle required to repress transposon activity during meiosis. Expressed in the cytoplasm of germline stem cells, cyst cells, nurse cells and oocytes, with strong accumulation in region IIb germline cysts in the germarium. In post-germarium egg chambers accumulates in the perinuclear loci that appear to be the nuage. With evidence from 7 publications.
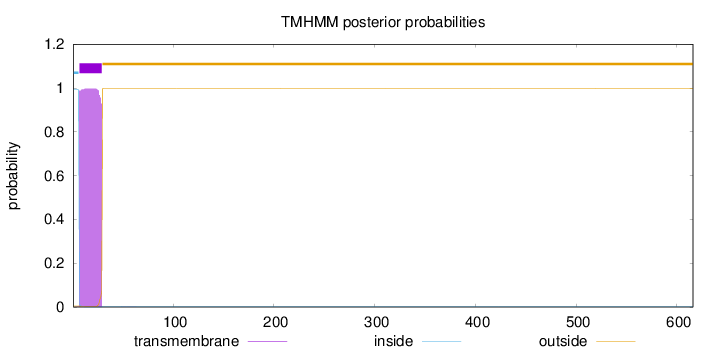
Length:
616
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.76228
Exp number, first 60 AAs:
22.76124
Total prob of N-in:
0.99496
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 616
Population Genetic Test Statistics
Pi
273.970206
Theta
174.082883
Tajima's D
1.598788
CLR
0.579402
CSRT
0.807609619519024
Interpretation
Uncertain
