Gene
KWMTBOMO01136
Pre Gene Modal
BGIBMGA007465
Annotation
PREDICTED:_2-phosphoxylose_phosphatase_1_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.278 Nuclear Reliability : 1.084
Sequence
CDS
ATGCCGCCTGTACTCACAAATAAAAACAGAATAATGAGTGAAATGATGAAGCTTTCATTTCATCACAGAGCTTTTTATTGTTACCTCGGCATGAGTCTCTGGATTTTTGTTCTCATAACGGTGATGTACAAATACATGTCGGTGAGCGAAGAAATAAGCATACCCAAGAAAATATATAAAGATCAATACATATCAAAAAGTGATGTTAAGTTTAGAAAGATATATATGAGAGCTTGCAATCCAGCTGATAATATAGTGAGAGGTTCAGAAGGTGCAGTAGATGAGGATGCTTGGTTGTTGCATGGTGTGCTCGTGATCACCAGGCATGGAGATAGGGGTCCACTAACACATCTCAAAGGAGGTGATAAATTACCTTGTGATTCGTCAATTGTTTCGCCACTGTTGAAAAGTTACGAAGACTATGTAGCTAACGCAAGCGCGTCAGGTCGGGCGTGGTGGGTGACCTCAACAGGTCCTTTTCACAATTTCCCTTCACTGCCGACTCGGGCGGCCGGGACCAGCTGTGCCCTGGGACAGTTGACGCCAAGGGGACTCCTGCAAATGATAACTGTGGGTAACATTCTGAGGGCAGCGTATGCAGATAAATTAGGATTAGACAATATTGATCCACAGGGAAAAAAGGACACAGGTGTGGCGGTGTCGTACAGCACCCGCTACCGCCGCACGTTCCTGTCGCTTTCGGGGGCGTGGTGGGGGGCGGCGCGGGGCGGGGCGGGGGCGCGGCTGGCTGCCTGTCGGGAGGCGCACAGCGTCTCCTTCTGCTTCAGGGACTGCGCCTGTCCCGCTCATCACGCCATCGACAAAAAAATCAACAATCAAGCAAGGAATCGCTTAGAGTCTCATCCCGCAATCAAAGAGCTGGTCAGCAAGTTATCTAAAGTGTTATTCGAGACGCAAGAACGTACAGACGCGGATGTCGTACGGGATGCCCTTCTTGCCTATATGTGCCACGATGTCCCTTTGCCGTGCACGCACAAAGCCAATCACGAAAAACCAAAACTGACCATAGACAAAAGGCACAGACTACACGAAAGCAAAACAGGAACCAGAAACCTCTTAGATGTCGACATCGACACTTTGAATATGGAACTGGATTATATAAACAATCAGGTGGATTTAAACGGAGACGTGGGTAGGAAAGCCAGAGAAGTCATCGGAAAATACTATGACCTGAAAGATAAGAAAGCGCCCTTAGATTTTGATGCCCAAATGGAGAGGGAGAAGTTGTTGTATTACCAGCAGAGATATTTGGATACCGGCGACACGTACGACGACGTAGTCATAGTGAAGAAGAATTTGGACGCGGATTTTAATTTTCCGAACGACGCTAGAATCGATGCGGACTTCGACGAAGACTACAAGGAGCCGACGCCCGAAACCGAAGACTTCTGCATCAAAAAAGAGCACGTTCTCTCGTTGTTCGCGTACTTGGAGTGGAGCTACCGGCAGGAGATCAAGAATATTCACAATCGGAAACGGGGCTTGCTCGTCTCGTACGGTCTGCTGCACAACATAGTCCAGAACATGATACGCATGATATCGGAGAACAAGCCCAGGTTCGTGATATACTCCGGTCACGATAAGACGTTACAGGCGCTCGTCTTGGCCCTCGGCTTGAGCAATTACCAATATTACAACATACAGTACGCCTCGAGATTGATATTTGAAGTGTACAGGAAGAAGGATCTGCGCGATGAGGTCAAATACGCGAAGAGAAAGGCCGTGGCCCGGGACTTTTACTTCCGGGTCGTCTTCAACGGCGAGGACGTGACGAACAAACTTAGTTTCTGCAACGCGAAACACAATGTCGTTATGAGGGTAGTCGATCCAATAGATGACTTGAAAATTTACAATACATTTTTGTGTCCGATCGAGAGCATCGTAAGGTTTATACACGACGATTATTTTAGTGTTTTTAATGTTAGTAACTTCAAGGATGCGTGCGCCACGGCCGGTAACAAGCGTGGATGA
Protein
MPPVLTNKNRIMSEMMKLSFHHRAFYCYLGMSLWIFVLITVMYKYMSVSEEISIPKKIYKDQYISKSDVKFRKIYMRACNPADNIVRGSEGAVDEDAWLLHGVLVITRHGDRGPLTHLKGGDKLPCDSSIVSPLLKSYEDYVANASASGRAWWVTSTGPFHNFPSLPTRAAGTSCALGQLTPRGLLQMITVGNILRAAYADKLGLDNIDPQGKKDTGVAVSYSTRYRRTFLSLSGAWWGAARGGAGARLAACREAHSVSFCFRDCACPAHHAIDKKINNQARNRLESHPAIKELVSKLSKVLFETQERTDADVVRDALLAYMCHDVPLPCTHKANHEKPKLTIDKRHRLHESKTGTRNLLDVDIDTLNMELDYINNQVDLNGDVGRKAREVIGKYYDLKDKKAPLDFDAQMEREKLLYYQQRYLDTGDTYDDVVIVKKNLDADFNFPNDARIDADFDEDYKEPTPETEDFCIKKEHVLSLFAYLEWSYRQEIKNIHNRKRGLLVSYGLLHNIVQNMIRMISENKPRFVIYSGHDKTLQALVLALGLSNYQYYNIQYASRLIFEVYRKKDLRDEVKYAKRKAVARDFYFRVVFNGEDVTNKLSFCNAKHNVVMRVVDPIDDLKIYNTFLCPIESIVRFIHDDYFSVFNVSNFKDACATAGNKRG
Summary
Similarity
Belongs to the histidine acid phosphatase family.
Uniprot
EMBL
BABH01036234
NWSH01004064
PCG65519.1
ODYU01008464
SOQ52095.1
KZ150100
+ More
PZC73531.1 KQ459232 KPJ02455.1 KQ460685 KPJ12809.1 AGBW02014038 OWR42360.1 GDHF01032382 GDHF01014933 JAI19932.1 JAI37381.1 GAKP01005544 JAC53408.1 CH963857 EDW75803.1 CH477971 EAT34717.1 CH379060 EAL33705.1 CH479180 EDW28891.1
PZC73531.1 KQ459232 KPJ02455.1 KQ460685 KPJ12809.1 AGBW02014038 OWR42360.1 GDHF01032382 GDHF01014933 JAI19932.1 JAI37381.1 GAKP01005544 JAC53408.1 CH963857 EDW75803.1 CH477971 EAT34717.1 CH379060 EAL33705.1 CH479180 EDW28891.1
Proteomes
PRIDE
Pfam
PF00328 His_Phos_2
SUPFAM
SSF53254
SSF53254
Gene 3D
ProteinModelPortal
Ontologies
KEGG
GO
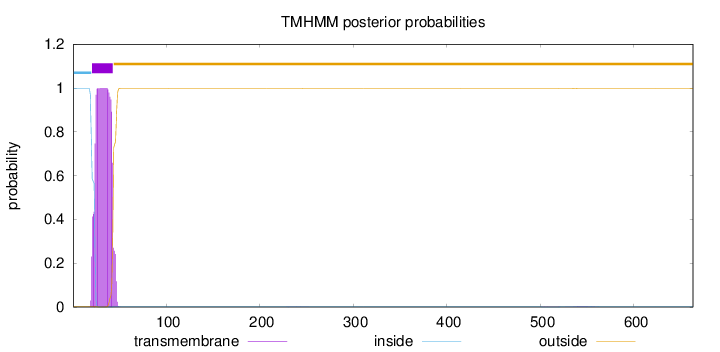
Topology
Length:
663
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.6553399999999
Exp number, first 60 AAs:
21.57574
Total prob of N-in:
0.99809
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 663
Population Genetic Test Statistics
Pi
231.932764
Theta
133.393697
Tajima's D
-2.422083
CLR
10.436519
CSRT
0.00169991500424979
Interpretation
Uncertain
