Gene
KWMTBOMO01134
Pre Gene Modal
BGIBMGA007467
Annotation
PREDICTED:_cyclin-dependent_kinase_5_activator_1_[Bombyx_mori]
Full name
Cyclin-dependent kinase 5 activator
Location in the cell
Mitochondrial Reliability : 1.21 Nuclear Reliability : 1.847
Sequence
CDS
ATGCGGGGACTTAGCGGCTGCATGCGCGCGGGCCTCATGGGCACCGTGCTGAGCTTCAGTCCGCGCGAGCGGCGACCGCCCGCCGCCGCGCCCCAACCCGATCGCGCCGCCCTCGCCTACTCCTACGAGCGTCTGAACAACGCCAAGAACAGGGAGAACCGCGAGCTGCCGCGGGACGAGCGACGCGCCACTCTCGACGATGCTGCCAAAATCATATCCGAGAAGACGGCCCTCGAAAAGAATCTCAAGAAGCATTCTCTGTTCATAAACGCTTTATCATGGAAGAGGTTCTCGACAGCGAACAATAACAAGAAGAAGCTAGAATCGAAGAGCAAGAGCGTGACGACATTCCGCGCGCCGCTGACGGACAATGCGCCGCTCGATCGCAACAAGAATGTTAGCGTGCAGGGGGGATACCCGCGGCCGGTGCCTGCGCCCCTACCGCCGCCTCCAGCTGAGGTGGTGCGAGCTAACGCACTCAACAACAATGTGTGCTACGCGGAGAAGCTGCCAGCTACGACGGTGGGGCCTGTGGTCGCACCGCGGAAGACCATAATCCAGGCCTCGACCTCAGAGCTGCTCAAATGCCTCGGCACGTTCTTGCATGCGCGGTGCCATCGCTTGCGTGACTTCCAAGCCGGCGACGCGGTCATGTGGCTGCGAACCGTCGATCGCTCTCTGCTTTTACAGGGCTGGCAGGACGTGGCATTCATAAACCCGGCGAACGTCGTGTTCGTGTACATGCTAGTCCGAGAGCTCGTGGACGGAGAAAGGATAGCGCGTCCTCAGGAGCTGCAGGCCGTCGTTCTCACCTGCCTCTACCTCTCTTACTCTTATATGGGCAACGAGATCTCGTACCCCCTCAAGCCGTTCCTAGTTGAAGACTCTAAAGACAAATTCTGGGACCGATGCCTCTTAATCGTCGATCGGCTATCGTTCAACATGCTGAGGATCAATTCGGAGCCGGGTTTCTTCACGGAAGTGTTCACGGAGCTGAAGGCGTGCGGCGTGGTGGACAGCCCTCCTCCCGCACCACCACACCCCGCCCCCGTACCCCCTCACACCGTCACGGCCGCCTGA
Protein
MRGLSGCMRAGLMGTVLSFSPRERRPPAAAPQPDRAALAYSYERLNNAKNRENRELPRDERRATLDDAAKIISEKTALEKNLKKHSLFINALSWKRFSTANNNKKKLESKSKSVTTFRAPLTDNAPLDRNKNVSVQGGYPRPVPAPLPPPPAEVVRANALNNNVCYAEKLPATTVGPVVAPRKTIIQASTSELLKCLGTFLHARCHRLRDFQAGDAVMWLRTVDRSLLLQGWQDVAFINPANVVFVYMLVRELVDGERIARPQELQAVVLTCLYLSYSYMGNEISYPLKPFLVEDSKDKFWDRCLLIVDRLSFNMLRINSEPGFFTEVFTELKACGVVDSPPPAPPHPAPVPPHTVTAA
Summary
Subunit
Heterodimer of a catalytic subunit and a regulatory subunit.
Similarity
Belongs to the cyclin-dependent kinase 5 activator family.
Uniprot
A0A2H1WTL2
A0A194QA94
A0A2W1B9Q2
A0A2A4JAT2
D6WWN6
A0A1W4XKY3
+ More
A0A1B6L2I7 A0A1Y1KRF3 A0A151J941 A0A3L8DZY4 V9IKL4 A0A088AVN4 A0A158P1T2 A0A2J7RQU1 A0A1B6C3W1 A0A151HZP7 K7IRJ6 N6TBR5 A0A026WPL3 A0A154PI09 A0A1A9ZQV9 B4KJK8 A0A1A9VJA2 A0A1B0BUT3 A0A3B0JLB3 B4G7J4 A0A1B0GAL8 B4MG57 J9K8D0 B3MPQ8 A0A2H8TIC5 A0A2S2PD93 A0A1B6HTQ0 B7P9I4 A0A2P6KIN4 A0A0A1XCR4 A0A0L8HF60 B4HWK3 E9H3I5 A0A210R1W6 A0A1A9YD76 V4ATH6 A0A2T7PBE4 C3ZR40 A0A2C9JLJ6 A0A0B6ZC47 A0A2D0SA81 A0A3B4DK52 A0A3P8S5L9 A0A1S3NGU3 F6SNC8 G3NI28 F1Q6W4 A0A3P8ZR02 A0A3B4YJU0 A0A3B4U772
A0A1B6L2I7 A0A1Y1KRF3 A0A151J941 A0A3L8DZY4 V9IKL4 A0A088AVN4 A0A158P1T2 A0A2J7RQU1 A0A1B6C3W1 A0A151HZP7 K7IRJ6 N6TBR5 A0A026WPL3 A0A154PI09 A0A1A9ZQV9 B4KJK8 A0A1A9VJA2 A0A1B0BUT3 A0A3B0JLB3 B4G7J4 A0A1B0GAL8 B4MG57 J9K8D0 B3MPQ8 A0A2H8TIC5 A0A2S2PD93 A0A1B6HTQ0 B7P9I4 A0A2P6KIN4 A0A0A1XCR4 A0A0L8HF60 B4HWK3 E9H3I5 A0A210R1W6 A0A1A9YD76 V4ATH6 A0A2T7PBE4 C3ZR40 A0A2C9JLJ6 A0A0B6ZC47 A0A2D0SA81 A0A3B4DK52 A0A3P8S5L9 A0A1S3NGU3 F6SNC8 G3NI28 F1Q6W4 A0A3P8ZR02 A0A3B4YJU0 A0A3B4U772
Pubmed
EMBL
ODYU01010904
SOQ56306.1
KQ459232
KPJ02453.1
KZ150241
PZC71908.1
+ More
NWSH01002253 PCG68808.1 KQ971361 EFA08111.1 GEBQ01022401 GEBQ01022035 JAT17576.1 JAT17942.1 GEZM01075893 JAV63894.1 KQ979492 KYN21211.1 QOIP01000002 RLU25967.1 JR049103 AEY60859.1 ADTU01006783 NEVH01000618 PNF43187.1 GEDC01029106 JAS08192.1 KQ976675 KYM78229.1 APGK01036344 KB740939 KB632013 ENN77729.1 ERL88023.1 KK107138 EZA57965.1 KQ434923 KZC11479.1 CH933807 EDW13588.2 KRG03888.1 JXJN01020906 JXJN01020907 JXJN01020908 OUUW01000006 SPP81633.1 CH479180 EDW28394.1 CCAG010011279 CH940671 EDW58195.1 ABLF02021288 ABLF02021291 CH902620 EDV32306.1 GFXV01001647 MBW13452.1 GGMR01014790 MBY27409.1 GECU01029675 JAS78031.1 ABJB010127749 ABJB010333436 DS664098 EEC03256.1 MWRG01011051 PRD26171.1 GBXI01005556 JAD08736.1 KQ418303 KOF87888.1 CH480818 EDW52398.1 GL732588 EFX73779.1 NEDP02000857 OWF54881.1 KB201360 ESO97041.1 PZQS01000005 PVD30744.1 GG666664 EEN44913.1 HACG01019304 CEK66169.1 AAMC01124167 KV460639 OCA16249.1 CR391936
NWSH01002253 PCG68808.1 KQ971361 EFA08111.1 GEBQ01022401 GEBQ01022035 JAT17576.1 JAT17942.1 GEZM01075893 JAV63894.1 KQ979492 KYN21211.1 QOIP01000002 RLU25967.1 JR049103 AEY60859.1 ADTU01006783 NEVH01000618 PNF43187.1 GEDC01029106 JAS08192.1 KQ976675 KYM78229.1 APGK01036344 KB740939 KB632013 ENN77729.1 ERL88023.1 KK107138 EZA57965.1 KQ434923 KZC11479.1 CH933807 EDW13588.2 KRG03888.1 JXJN01020906 JXJN01020907 JXJN01020908 OUUW01000006 SPP81633.1 CH479180 EDW28394.1 CCAG010011279 CH940671 EDW58195.1 ABLF02021288 ABLF02021291 CH902620 EDV32306.1 GFXV01001647 MBW13452.1 GGMR01014790 MBY27409.1 GECU01029675 JAS78031.1 ABJB010127749 ABJB010333436 DS664098 EEC03256.1 MWRG01011051 PRD26171.1 GBXI01005556 JAD08736.1 KQ418303 KOF87888.1 CH480818 EDW52398.1 GL732588 EFX73779.1 NEDP02000857 OWF54881.1 KB201360 ESO97041.1 PZQS01000005 PVD30744.1 GG666664 EEN44913.1 HACG01019304 CEK66169.1 AAMC01124167 KV460639 OCA16249.1 CR391936
Proteomes
UP000053268
UP000218220
UP000007266
UP000192223
UP000078492
UP000279307
+ More
UP000005203 UP000005205 UP000235965 UP000078540 UP000002358 UP000019118 UP000030742 UP000053097 UP000076502 UP000092445 UP000009192 UP000078200 UP000092460 UP000268350 UP000008744 UP000092444 UP000008792 UP000007819 UP000007801 UP000001555 UP000053454 UP000001292 UP000000305 UP000242188 UP000092443 UP000030746 UP000245119 UP000001554 UP000076420 UP000221080 UP000261440 UP000265080 UP000087266 UP000008143 UP000007635 UP000000437 UP000265140 UP000261360 UP000261420
UP000005203 UP000005205 UP000235965 UP000078540 UP000002358 UP000019118 UP000030742 UP000053097 UP000076502 UP000092445 UP000009192 UP000078200 UP000092460 UP000268350 UP000008744 UP000092444 UP000008792 UP000007819 UP000007801 UP000001555 UP000053454 UP000001292 UP000000305 UP000242188 UP000092443 UP000030746 UP000245119 UP000001554 UP000076420 UP000221080 UP000261440 UP000265080 UP000087266 UP000008143 UP000007635 UP000000437 UP000265140 UP000261360 UP000261420
PRIDE
Interpro
SUPFAM
SSF47954
SSF47954
CDD
ProteinModelPortal
A0A2H1WTL2
A0A194QA94
A0A2W1B9Q2
A0A2A4JAT2
D6WWN6
A0A1W4XKY3
+ More
A0A1B6L2I7 A0A1Y1KRF3 A0A151J941 A0A3L8DZY4 V9IKL4 A0A088AVN4 A0A158P1T2 A0A2J7RQU1 A0A1B6C3W1 A0A151HZP7 K7IRJ6 N6TBR5 A0A026WPL3 A0A154PI09 A0A1A9ZQV9 B4KJK8 A0A1A9VJA2 A0A1B0BUT3 A0A3B0JLB3 B4G7J4 A0A1B0GAL8 B4MG57 J9K8D0 B3MPQ8 A0A2H8TIC5 A0A2S2PD93 A0A1B6HTQ0 B7P9I4 A0A2P6KIN4 A0A0A1XCR4 A0A0L8HF60 B4HWK3 E9H3I5 A0A210R1W6 A0A1A9YD76 V4ATH6 A0A2T7PBE4 C3ZR40 A0A2C9JLJ6 A0A0B6ZC47 A0A2D0SA81 A0A3B4DK52 A0A3P8S5L9 A0A1S3NGU3 F6SNC8 G3NI28 F1Q6W4 A0A3P8ZR02 A0A3B4YJU0 A0A3B4U772
A0A1B6L2I7 A0A1Y1KRF3 A0A151J941 A0A3L8DZY4 V9IKL4 A0A088AVN4 A0A158P1T2 A0A2J7RQU1 A0A1B6C3W1 A0A151HZP7 K7IRJ6 N6TBR5 A0A026WPL3 A0A154PI09 A0A1A9ZQV9 B4KJK8 A0A1A9VJA2 A0A1B0BUT3 A0A3B0JLB3 B4G7J4 A0A1B0GAL8 B4MG57 J9K8D0 B3MPQ8 A0A2H8TIC5 A0A2S2PD93 A0A1B6HTQ0 B7P9I4 A0A2P6KIN4 A0A0A1XCR4 A0A0L8HF60 B4HWK3 E9H3I5 A0A210R1W6 A0A1A9YD76 V4ATH6 A0A2T7PBE4 C3ZR40 A0A2C9JLJ6 A0A0B6ZC47 A0A2D0SA81 A0A3B4DK52 A0A3P8S5L9 A0A1S3NGU3 F6SNC8 G3NI28 F1Q6W4 A0A3P8ZR02 A0A3B4YJU0 A0A3B4U772
PDB
1UNL
E-value=1.79893e-57,
Score=563
Ontologies
GO
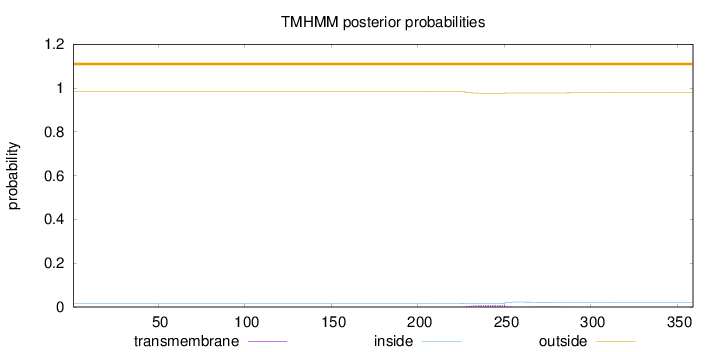
Topology
Length:
359
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.22016
Exp number, first 60 AAs:
0.00106
Total prob of N-in:
0.01693
outside
1 - 359
Population Genetic Test Statistics
Pi
274.350909
Theta
194.149402
Tajima's D
0.53831
CLR
2.566969
CSRT
0.536973151342433
Interpretation
Uncertain
