Gene
KWMTBOMO01133
Pre Gene Modal
BGIBMGA007468
Annotation
PREDICTED:_chromosome_transmission_fidelity_protein_18_homolog_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.522
Sequence
CDS
ATGAGTGACTATCCAGATCCGGACGAAGAATATGAACTTATGTACGCTGATGAACTGGAATTGATGCAAGAACAAGATGCTCTAACAGACACTAATGTAGTTGCAAAATCAAAAATTGCCAAAAGAAGCCTAGATTTTTCAAATCACATGTCAAGCCAACCAGCACCATCTACTCTCATAACAAAAACTACAAGTAGTACTGAAACACCTGTAACACCAAACGAAGATGCTGATACCGAAACCATCATCAATAGTCAGTCTTCAGTCATATTGCAACCAATGGAAGAGGCCTTGGTGTCCAATAAACGTACTGCAGATGAATTGTTCGGCGATATTGAAGATATAGACTTTGATGATAAAGAAATGCCAAACAAAAGATTTAAGAGTGAAGAGGAAAATGATTTAGAATTAATAAACAAGATTATAGAGGGAAGGAGGTTGCGGCAGTTGCTAGTGGAACCAAGTAATAGACTCCAGGGAACAAATAAACCCAAGTATGATGTGAAAGGAAATATGTCTTACAATATACCAAGGTGGGCTTTTGTACCTCTAACAAATTTTGAAGGTGACAGAATATACATCAGGTTAGAATCTGAAGAATCATGGGAAAATAATCTAAATATTAATATAGAACAGCCATCTCTACATGCAATGTATGCTAATGCTTGGGAGGAAGCTAGGAAAATTTTAGAAAAAAAACAAGAGCGTCTAATAGAAAACGAAAATACAGAAGAAGTGGTAACTATTGAAGAAGATATGGAGACAAAAGATGCACAGCTATGGGTGGATAAGTACAGGCCCCAGTCGTATGTAGATCTATTATCTGAAGAACCGGTAAACAGAGCTCTATTACACTGGTTACATTTATGGGATAAGATCGTTTTCAAAAAAGAAGTCAAAACGAAAGACCCCCAACAGCGGAACAGCTTCGTCAAGCGCACGGGACAGTTCCAACTGTTCAACAAATGGAAAGGAAAGACGGATAACGAGCCCGAGTTAGACGAAGACGGAAGACCTCATCACAAGGTGGCGCTGCTGTGCGGCCCTCCCGGAGTGGGGAAGACGACCCTGGCACATTTATTAGCTAAACTAGCCGGTTACCGACCGGTAGAAATAAACGCATCTGATGAGCGAAGTACGGACGCGTTCCGGACAGCCCTGCAAAGCGCCACGCTTATGAGATCTGTGCTGGACGCCGAGAAAAGACCGAACTGTTTGATATTAGATGAAATAGATGGTGCCCCCTTGCCGACGATCGAGTTGTTAGTGAAGTGGTGTACGGCAGCCATCGACAGCGGGAAGAAGAAGACCAAATCGCAGCCTCTGAGACGTCCCATTATTGCTATATGTAATGACTTGTACGCTACATCATTGAGGCCTTTGAGAGCGGTGGCGCTGTTGGTGCGCGTGGGCGGCGCGTCAGTGTGCCGGCTGTCGGCGCGGCTGGCGGCCGTGTGCGCGCGCGAGGGGCTGCTGGCGGCGCCGCACGTGCTCACCGCGCTCGCCACGCGCGCCCAGGGCGACGTCCGAGCCGCCATACAAGTTCTCAGCTTCATTAAAGCCAGAGCTAGAGCACAGTTAAAGGTAGAGGACGTTGAGAACGCAGCCATTGGCACCAAGGATTGCAACAAAACATTAATGCAAGCGTTACAAGCGGTATTCACGATCGACGACAAAGAACCGGAGGCCATATTGAAGATTGTACAGGCAGCCGGGGAGTATGACAGAATAGCGGACGGAATATTCGAGAATTACATGGCCGCCCGCGTGGACGCGCGGATGCTGATAGCTTGCGAGACTCTATCGTGGCTGCGCTTACTGGACGCCATTAGCTCGTGGACGATGAGGACCCAGAACTATTCGTTGTACGGGGCCCTGTCGTTATGTATAGCCAGGTGTCACAGGGTACTCGCATCGAGGGCCCCGCAAAGGGTCAAATTTCCTACACAGGCACAAGAGGCCTACAGAAAGAAATTAGAATTGGAGAGTATAGTGAATTCGGTATGGCGAGGCAGCGTGCCGTCAGTGGCACTTTCTAGAAACTCTTTGAAGGTTGACGTGTTACCGTTGCTGCCCTATCTGTTGTCGCCGTCACTACGCACCGCTAACGTACAGTTGTGCAGCGAGAGCGAGAGGAGGTCGCTGGGCGCATGCGCGGGCGCGCTGTGCGACTACGGGCTGCGGTACGTGCAGCAGCGGACCGCCGACGGACTCTACGCTTTCGTTTTGGAACCGGACATTAACAGATTAGCGTTTTACGGTAACGAAGACAGAGTCCGTCCGCCGCCGGCCGTCAGACAAGCCATCGCCCGGGCACAGCAGCTAGAAATCATACGAAGGAACGAGGAAATGACTGGTCGAGCTCCAAGCACAACTACATCAGATAGTAAAAAGCCAGTGGAGTCCAAGAAGCCAGCCACGACAATGAGGGAGCAGGACGCCGCTATACCCAACCACATGCAGCGCTTGAAACCGAAAGAAGTTACGAAAAGCACACCCCAGGTACACAAAGACTTCTTCGGTCGTATAGTTCCGCTGTCACAAGTCAAGCGACAAGATGCTTTGCCGGATGTTATATCTTCCAGCAGTGTATTCTACCAGTACAGAGAAGGCTTCAACAACGCGGTCAGAAGAAAAGTTCGCCTCCGTGATTTGCTGTAG
Protein
MSDYPDPDEEYELMYADELELMQEQDALTDTNVVAKSKIAKRSLDFSNHMSSQPAPSTLITKTTSSTETPVTPNEDADTETIINSQSSVILQPMEEALVSNKRTADELFGDIEDIDFDDKEMPNKRFKSEEENDLELINKIIEGRRLRQLLVEPSNRLQGTNKPKYDVKGNMSYNIPRWAFVPLTNFEGDRIYIRLESEESWENNLNINIEQPSLHAMYANAWEEARKILEKKQERLIENENTEEVVTIEEDMETKDAQLWVDKYRPQSYVDLLSEEPVNRALLHWLHLWDKIVFKKEVKTKDPQQRNSFVKRTGQFQLFNKWKGKTDNEPELDEDGRPHHKVALLCGPPGVGKTTLAHLLAKLAGYRPVEINASDERSTDAFRTALQSATLMRSVLDAEKRPNCLILDEIDGAPLPTIELLVKWCTAAIDSGKKKTKSQPLRRPIIAICNDLYATSLRPLRAVALLVRVGGASVCRLSARLAAVCAREGLLAAPHVLTALATRAQGDVRAAIQVLSFIKARARAQLKVEDVENAAIGTKDCNKTLMQALQAVFTIDDKEPEAILKIVQAAGEYDRIADGIFENYMAARVDARMLIACETLSWLRLLDAISSWTMRTQNYSLYGALSLCIARCHRVLASRAPQRVKFPTQAQEAYRKKLELESIVNSVWRGSVPSVALSRNSLKVDVLPLLPYLLSPSLRTANVQLCSESERRSLGACAGALCDYGLRYVQQRTADGLYAFVLEPDINRLAFYGNEDRVRPPPAVRQAIARAQQLEIIRRNEEMTGRAPSTTTSDSKKPVESKKPATTMREQDAAIPNHMQRLKPKEVTKSTPQVHKDFFGRIVPLSQVKRQDALPDVISSSSVFYQYREGFNNAVRRKVRLRDLL
Summary
Uniprot
H9JD71
A0A2A4JBK0
A0A2H1WQI1
A0A194QBF0
A0A194RAB7
A0A1B6LVK8
+ More
A0A1S4FJ24 A0A1B6J544 Q16Z43 A0A1B6HXM3 A0A1B6IFT1 A0A1Q3G079 A0A1Q3G0A2 A0A2S2QED8 A0A1I8PLA4 D6WXA2 A0A034VCE5 A0A1I8N525 B4KJY8 A0A2H8TMV2 B4NYE6 A0A0K8VP61 A0A0M4E7S0 A0A1B6D4D8 Q16Z42 Q8T3K3 A0A2S2NZK5 B4JQJ0 B4LRR8 A0A0A9W048 A0A1W4V823 Q95WV5 B4MVB5 Q8IQ05 B3N370 B4I327 X1WIE2 A0A0J9QV88 A0A3B0JUC2 U4UF04 B3MP61 A0A1Y1N551 Q29ME9 W8BNZ8 A0A182JLN0 A0A0T6B2L9 A0A182QTV9 A0A1A9ZQR2 A0A182VU88 A0A084VYL9 B4Q9P5 A0A023F1H3 A0A182VK68 A0A0L0BZE7 A0A1W4WCS3 A0A1S4H083 A0A182Y947 Q7PWS6 A0A182F5Q6 A0A336M1C1 N6UB29 A0A1J1IZN7 A0A1A9V0X3 A0A232FD56 A0A182RUR9 A0A182LN03 A0A182K417 A0A1S4FJ93 K7ISX5 A0A182MWD8 A0A088AC10 A0A0M9A6R5 A0A182P692 A0A131YFD6 A0A224YQ95 K1PLR8 A0A026W7S8 A0A293M885 A0A182I758 E2C5Y3 A0A1E1XDK9 H2LY12 A0A0L8GP69 V4AVS1 A0A3P8N8B2 A0A3P9C0F4 A0A0L8GPM4 A0A3P9JAE9 A0A3P9KU70 B7QM52 I3J5K6 A0A3B3VR70 A0A0L7QPR2 A0A3B3B7D1 A0A087Y8Y8 A0A210QQS4 V5IDU9 A0A1W5BEK2
A0A1S4FJ24 A0A1B6J544 Q16Z43 A0A1B6HXM3 A0A1B6IFT1 A0A1Q3G079 A0A1Q3G0A2 A0A2S2QED8 A0A1I8PLA4 D6WXA2 A0A034VCE5 A0A1I8N525 B4KJY8 A0A2H8TMV2 B4NYE6 A0A0K8VP61 A0A0M4E7S0 A0A1B6D4D8 Q16Z42 Q8T3K3 A0A2S2NZK5 B4JQJ0 B4LRR8 A0A0A9W048 A0A1W4V823 Q95WV5 B4MVB5 Q8IQ05 B3N370 B4I327 X1WIE2 A0A0J9QV88 A0A3B0JUC2 U4UF04 B3MP61 A0A1Y1N551 Q29ME9 W8BNZ8 A0A182JLN0 A0A0T6B2L9 A0A182QTV9 A0A1A9ZQR2 A0A182VU88 A0A084VYL9 B4Q9P5 A0A023F1H3 A0A182VK68 A0A0L0BZE7 A0A1W4WCS3 A0A1S4H083 A0A182Y947 Q7PWS6 A0A182F5Q6 A0A336M1C1 N6UB29 A0A1J1IZN7 A0A1A9V0X3 A0A232FD56 A0A182RUR9 A0A182LN03 A0A182K417 A0A1S4FJ93 K7ISX5 A0A182MWD8 A0A088AC10 A0A0M9A6R5 A0A182P692 A0A131YFD6 A0A224YQ95 K1PLR8 A0A026W7S8 A0A293M885 A0A182I758 E2C5Y3 A0A1E1XDK9 H2LY12 A0A0L8GP69 V4AVS1 A0A3P8N8B2 A0A3P9C0F4 A0A0L8GPM4 A0A3P9JAE9 A0A3P9KU70 B7QM52 I3J5K6 A0A3B3VR70 A0A0L7QPR2 A0A3B3B7D1 A0A087Y8Y8 A0A210QQS4 V5IDU9 A0A1W5BEK2
Pubmed
19121390
26354079
17510324
18362917
19820115
25348373
+ More
25315136 17994087 17550304 25401762 11161570 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 23537049 28004739 15632085 24495485 24438588 25474469 26108605 12364791 25244985 28648823 20966253 20075255 26830274 28797301 22992520 24508170 20798317 28503490 17554307 23254933 25186727 29451363 28812685 25765539
25315136 17994087 17550304 25401762 11161570 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 23537049 28004739 15632085 24495485 24438588 25474469 26108605 12364791 25244985 28648823 20966253 20075255 26830274 28797301 22992520 24508170 20798317 28503490 17554307 23254933 25186727 29451363 28812685 25765539
EMBL
BABH01036228
BABH01036229
NWSH01002253
PCG68810.1
ODYU01010268
SOQ55257.1
+ More
KQ459249 KPJ02315.1 KQ460685 KPJ12806.1 GEBQ01012255 JAT27722.1 GECU01013445 JAS94261.1 CH477499 EAT39908.1 GECU01028297 JAS79409.1 GECU01021928 JAS85778.1 GFDL01001836 JAV33209.1 GFDL01001839 JAV33206.1 GGMS01006850 MBY76053.1 KQ971361 EFA08812.2 GAKP01019764 JAC39188.1 CH933807 EDW12591.1 GFXV01003688 MBW15493.1 CM000157 EDW87579.1 GDHF01011969 JAI40345.1 CP012523 ALC38865.1 GEDC01026847 GEDC01016751 JAS10451.1 JAS20547.1 EAT39909.1 AY094968 AAM11321.1 GGMR01009517 MBY22136.1 CH916372 EDV99170.1 CH940649 EDW64670.1 GBHO01045364 GBHO01045361 GBHO01045360 GBHO01045359 JAF98239.1 JAF98242.1 JAF98243.1 JAF98244.1 AF287273 AAL02426.1 CH963857 EDW76460.1 AE014134 AAN10380.2 CH954177 EDV57669.1 CH480820 EDW54172.1 ABLF02036818 ABLF02036821 CM002910 KMY87958.1 OUUW01000004 SPP79070.1 KB632064 ERL88495.1 CH902620 EDV31227.2 GEZM01014729 JAV92020.1 CH379060 EAL33745.2 GAMC01003690 JAC02866.1 LJIG01016173 KRT81397.1 AXCN02002086 ATLV01018378 KE525231 KFB43063.1 CM000361 EDX03656.1 GBBI01003851 JAC14861.1 JRES01001205 KNC24619.1 AAAB01008984 EAA14898.4 UFQS01000240 UFQT01000240 SSX01870.1 SSX22247.1 APGK01035573 APGK01035574 KB740928 ENN77861.1 CVRI01000064 CRL05020.1 NNAY01000433 OXU28423.1 AAZX01004589 AXCM01001391 KQ435724 KOX78380.1 GEDV01010513 JAP78044.1 GFPF01006783 MAA17929.1 JH818838 EKC19774.1 KK107348 EZA52142.1 GFWV01012363 MAA37092.1 APCN01003367 GL452875 EFN76642.1 GFAC01001846 JAT97342.1 KQ420929 KOF78818.1 KB200521 ESP01463.1 KOF78817.1 ABJB010436061 ABJB010537462 ABJB010566634 DS969253 EEC19924.1 AERX01039553 AERX01039554 KQ414806 KOC60628.1 AYCK01023044 AYCK01023045 AYCK01023046 NEDP02002358 OWF51090.1 GANP01011307 JAB73161.1
KQ459249 KPJ02315.1 KQ460685 KPJ12806.1 GEBQ01012255 JAT27722.1 GECU01013445 JAS94261.1 CH477499 EAT39908.1 GECU01028297 JAS79409.1 GECU01021928 JAS85778.1 GFDL01001836 JAV33209.1 GFDL01001839 JAV33206.1 GGMS01006850 MBY76053.1 KQ971361 EFA08812.2 GAKP01019764 JAC39188.1 CH933807 EDW12591.1 GFXV01003688 MBW15493.1 CM000157 EDW87579.1 GDHF01011969 JAI40345.1 CP012523 ALC38865.1 GEDC01026847 GEDC01016751 JAS10451.1 JAS20547.1 EAT39909.1 AY094968 AAM11321.1 GGMR01009517 MBY22136.1 CH916372 EDV99170.1 CH940649 EDW64670.1 GBHO01045364 GBHO01045361 GBHO01045360 GBHO01045359 JAF98239.1 JAF98242.1 JAF98243.1 JAF98244.1 AF287273 AAL02426.1 CH963857 EDW76460.1 AE014134 AAN10380.2 CH954177 EDV57669.1 CH480820 EDW54172.1 ABLF02036818 ABLF02036821 CM002910 KMY87958.1 OUUW01000004 SPP79070.1 KB632064 ERL88495.1 CH902620 EDV31227.2 GEZM01014729 JAV92020.1 CH379060 EAL33745.2 GAMC01003690 JAC02866.1 LJIG01016173 KRT81397.1 AXCN02002086 ATLV01018378 KE525231 KFB43063.1 CM000361 EDX03656.1 GBBI01003851 JAC14861.1 JRES01001205 KNC24619.1 AAAB01008984 EAA14898.4 UFQS01000240 UFQT01000240 SSX01870.1 SSX22247.1 APGK01035573 APGK01035574 KB740928 ENN77861.1 CVRI01000064 CRL05020.1 NNAY01000433 OXU28423.1 AAZX01004589 AXCM01001391 KQ435724 KOX78380.1 GEDV01010513 JAP78044.1 GFPF01006783 MAA17929.1 JH818838 EKC19774.1 KK107348 EZA52142.1 GFWV01012363 MAA37092.1 APCN01003367 GL452875 EFN76642.1 GFAC01001846 JAT97342.1 KQ420929 KOF78818.1 KB200521 ESP01463.1 KOF78817.1 ABJB010436061 ABJB010537462 ABJB010566634 DS969253 EEC19924.1 AERX01039553 AERX01039554 KQ414806 KOC60628.1 AYCK01023044 AYCK01023045 AYCK01023046 NEDP02002358 OWF51090.1 GANP01011307 JAB73161.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000008820
UP000095300
+ More
UP000007266 UP000095301 UP000009192 UP000002282 UP000092553 UP000001070 UP000008792 UP000192221 UP000007798 UP000000803 UP000008711 UP000001292 UP000007819 UP000268350 UP000030742 UP000007801 UP000001819 UP000075880 UP000075886 UP000092445 UP000075920 UP000030765 UP000000304 UP000075903 UP000037069 UP000192223 UP000076408 UP000007062 UP000069272 UP000019118 UP000183832 UP000078200 UP000215335 UP000075900 UP000075882 UP000075881 UP000002358 UP000075883 UP000005203 UP000053105 UP000075885 UP000005408 UP000053097 UP000075840 UP000008237 UP000001038 UP000053454 UP000030746 UP000265100 UP000265160 UP000265200 UP000265180 UP000001555 UP000005207 UP000261500 UP000053825 UP000261560 UP000028760 UP000242188
UP000007266 UP000095301 UP000009192 UP000002282 UP000092553 UP000001070 UP000008792 UP000192221 UP000007798 UP000000803 UP000008711 UP000001292 UP000007819 UP000268350 UP000030742 UP000007801 UP000001819 UP000075880 UP000075886 UP000092445 UP000075920 UP000030765 UP000000304 UP000075903 UP000037069 UP000192223 UP000076408 UP000007062 UP000069272 UP000019118 UP000183832 UP000078200 UP000215335 UP000075900 UP000075882 UP000075881 UP000002358 UP000075883 UP000005203 UP000053105 UP000075885 UP000005408 UP000053097 UP000075840 UP000008237 UP000001038 UP000053454 UP000030746 UP000265100 UP000265160 UP000265200 UP000265180 UP000001555 UP000005207 UP000261500 UP000053825 UP000261560 UP000028760 UP000242188
Interpro
IPR027417
P-loop_NTPase
+ More
IPR003959 ATPase_AAA_core
IPR003593 AAA+_ATPase
IPR013725 DNA_replication_fac_RFC1_C
IPR036273 CRAL/TRIO_N_dom_sf
IPR036865 CRAL-TRIO_dom_sf
IPR036880 Kunitz_BPTI_sf
IPR002223 Kunitz_BPTI
IPR001251 CRAL-TRIO_dom
IPR020843 PKS_ER
IPR013154 ADH_N
IPR011032 GroES-like_sf
IPR036291 NAD(P)-bd_dom_sf
IPR003959 ATPase_AAA_core
IPR003593 AAA+_ATPase
IPR013725 DNA_replication_fac_RFC1_C
IPR036273 CRAL/TRIO_N_dom_sf
IPR036865 CRAL-TRIO_dom_sf
IPR036880 Kunitz_BPTI_sf
IPR002223 Kunitz_BPTI
IPR001251 CRAL-TRIO_dom
IPR020843 PKS_ER
IPR013154 ADH_N
IPR011032 GroES-like_sf
IPR036291 NAD(P)-bd_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JD71
A0A2A4JBK0
A0A2H1WQI1
A0A194QBF0
A0A194RAB7
A0A1B6LVK8
+ More
A0A1S4FJ24 A0A1B6J544 Q16Z43 A0A1B6HXM3 A0A1B6IFT1 A0A1Q3G079 A0A1Q3G0A2 A0A2S2QED8 A0A1I8PLA4 D6WXA2 A0A034VCE5 A0A1I8N525 B4KJY8 A0A2H8TMV2 B4NYE6 A0A0K8VP61 A0A0M4E7S0 A0A1B6D4D8 Q16Z42 Q8T3K3 A0A2S2NZK5 B4JQJ0 B4LRR8 A0A0A9W048 A0A1W4V823 Q95WV5 B4MVB5 Q8IQ05 B3N370 B4I327 X1WIE2 A0A0J9QV88 A0A3B0JUC2 U4UF04 B3MP61 A0A1Y1N551 Q29ME9 W8BNZ8 A0A182JLN0 A0A0T6B2L9 A0A182QTV9 A0A1A9ZQR2 A0A182VU88 A0A084VYL9 B4Q9P5 A0A023F1H3 A0A182VK68 A0A0L0BZE7 A0A1W4WCS3 A0A1S4H083 A0A182Y947 Q7PWS6 A0A182F5Q6 A0A336M1C1 N6UB29 A0A1J1IZN7 A0A1A9V0X3 A0A232FD56 A0A182RUR9 A0A182LN03 A0A182K417 A0A1S4FJ93 K7ISX5 A0A182MWD8 A0A088AC10 A0A0M9A6R5 A0A182P692 A0A131YFD6 A0A224YQ95 K1PLR8 A0A026W7S8 A0A293M885 A0A182I758 E2C5Y3 A0A1E1XDK9 H2LY12 A0A0L8GP69 V4AVS1 A0A3P8N8B2 A0A3P9C0F4 A0A0L8GPM4 A0A3P9JAE9 A0A3P9KU70 B7QM52 I3J5K6 A0A3B3VR70 A0A0L7QPR2 A0A3B3B7D1 A0A087Y8Y8 A0A210QQS4 V5IDU9 A0A1W5BEK2
A0A1S4FJ24 A0A1B6J544 Q16Z43 A0A1B6HXM3 A0A1B6IFT1 A0A1Q3G079 A0A1Q3G0A2 A0A2S2QED8 A0A1I8PLA4 D6WXA2 A0A034VCE5 A0A1I8N525 B4KJY8 A0A2H8TMV2 B4NYE6 A0A0K8VP61 A0A0M4E7S0 A0A1B6D4D8 Q16Z42 Q8T3K3 A0A2S2NZK5 B4JQJ0 B4LRR8 A0A0A9W048 A0A1W4V823 Q95WV5 B4MVB5 Q8IQ05 B3N370 B4I327 X1WIE2 A0A0J9QV88 A0A3B0JUC2 U4UF04 B3MP61 A0A1Y1N551 Q29ME9 W8BNZ8 A0A182JLN0 A0A0T6B2L9 A0A182QTV9 A0A1A9ZQR2 A0A182VU88 A0A084VYL9 B4Q9P5 A0A023F1H3 A0A182VK68 A0A0L0BZE7 A0A1W4WCS3 A0A1S4H083 A0A182Y947 Q7PWS6 A0A182F5Q6 A0A336M1C1 N6UB29 A0A1J1IZN7 A0A1A9V0X3 A0A232FD56 A0A182RUR9 A0A182LN03 A0A182K417 A0A1S4FJ93 K7ISX5 A0A182MWD8 A0A088AC10 A0A0M9A6R5 A0A182P692 A0A131YFD6 A0A224YQ95 K1PLR8 A0A026W7S8 A0A293M885 A0A182I758 E2C5Y3 A0A1E1XDK9 H2LY12 A0A0L8GP69 V4AVS1 A0A3P8N8B2 A0A3P9C0F4 A0A0L8GPM4 A0A3P9JAE9 A0A3P9KU70 B7QM52 I3J5K6 A0A3B3VR70 A0A0L7QPR2 A0A3B3B7D1 A0A087Y8Y8 A0A210QQS4 V5IDU9 A0A1W5BEK2
PDB
1SXJ
E-value=8.30709e-10,
Score=156
Ontologies
GO
Topology
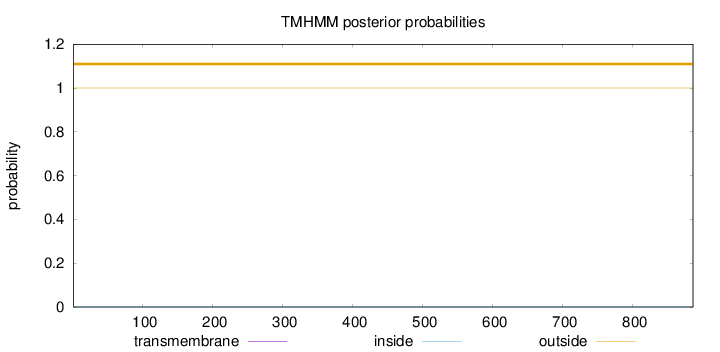
Length:
886
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00318
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 886
Population Genetic Test Statistics
Pi
200.550011
Theta
174.175488
Tajima's D
0.619442
CLR
0.468277
CSRT
0.543772811359432
Interpretation
Uncertain
