Gene
KWMTBOMO01129 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009430
Annotation
F-actin_capping_protein_beta_subunit_[Bombyx_mori]
Full name
F-actin-capping protein subunit beta
+ More
F-actin-capping protein subunit beta isoforms 1 and 2
F-actin-capping protein subunit beta isoforms 1 and 2
Alternative Name
Beta-actinin subunit II
CapZ 36/32
CapZ B1 and B2
CapZ beta
CapZ 36/32
CapZ B1 and B2
CapZ beta
Location in the cell
Nuclear Reliability : 3.008
Sequence
CDS
ATGAGTGACCAACAAATGGATTGTGCACTGGACTTGATGCGGAGGCTGCCCCCTCAGCAGATCGAAAAGAATTTAACAGATTTGATAGATCTCGTTCCGAGTATGTGTGACGACCTATTATCATCAGTAGATCAGCCTCTAAAGATTGCTCAGGATCGTAGCAACGGGAAAGACTATTTATTATGCGATTACAATCGCGACGGTGACTCCTATAGGTCTCCTTGGTCGAATACTTACGACCCACCATTAGATGATGGCTCCATGCCCTCTGAACGCTTGAGAAAACTAGAAATAGATGCCAACCTCGCCTTTGATCAATATCGAGAGATGTACTTTGAAGGTGGCGTTAGCTCAGTCTACCTTTGGGATATGGATCATGGCTTTGCAGGAGTAATATTAATCAAGAAAGCTGGAGATGGTTCCCAAAAGATCAAAGGATGCTGGGATTCAATCCACGTAGTGGAGGTGATCGAGAAGAGTTCAGGACGCAATGCTCACTACAAGTTGACCTCGACTGCAATGTTGTGGCTTCAGACTAATAAAGAAAGCAGCGGCACGATGAACCTTGGAGGCAGTTTGACTAGACAGGCAGAACAAGACTCGACAGTAAGTGATGTGACTCCGCACATTGCAAATATTGGGCGCATGGTGGAAGACATGGAAAATAAGATCAGAAACACACTTAACGACATTTATTTTGGTAAAACAAAAGATATAGTGAGCGGGCTGAGGTCAGTGATCCCGGCGGACGTGGCGCGCCGCACCGCCGCGCTGCAGCACGACCTCGCGCTCGCCCTGCAGCGCCGCCACGTGCAGCGCGACGACTGA
Protein
MSDQQMDCALDLMRRLPPQQIEKNLTDLIDLVPSMCDDLLSSVDQPLKIAQDRSNGKDYLLCDYNRDGDSYRSPWSNTYDPPLDDGSMPSERLRKLEIDANLAFDQYREMYFEGGVSSVYLWDMDHGFAGVILIKKAGDGSQKIKGCWDSIHVVEVIEKSSGRNAHYKLTSTAMLWLQTNKESSGTMNLGGSLTRQAEQDSTVSDVTPHIANIGRMVEDMENKIRNTLNDIYFGKTKDIVSGLRSVIPADVARRTAALQHDLALALQRRHVQRDD
Summary
Description
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments.
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments. Isoform 3 may play a role in spermatogenesis. Alternatively, may play a role in later maturation steps such as capacitation and fertilization which involve changes of membrane domains. Plays a role in the regulation of cell morphology and cytoskeletal organization (By similarity).
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments. Plays a role in the regulation of cell morphology and cytoskeletal organization.
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments. Plays a role in the regulation of cell morphology and cytoskeletal organization (By similarity).
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments. Isoform 3 may play a role in spermatogenesis. Alternatively, may play a role in later maturation steps such as capacitation and fertilization which involve changes of membrane domains. Plays a role in the regulation of cell morphology and cytoskeletal organization (By similarity).
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments. Plays a role in the regulation of cell morphology and cytoskeletal organization.
F-actin-capping proteins bind in a Ca(2+)-independent manner to the fast growing ends of actin filaments (barbed end) thereby blocking the exchange of subunits at these ends. Unlike other capping proteins (such as gelsolin and severin), these proteins do not sever actin filaments. Plays a role in the regulation of cell morphology and cytoskeletal organization (By similarity).
Subunit
Heterodimer of an alpha and a beta subunit.
Heterodimer of an alpha and a beta subunit. Component of the WASH complex (By similarity). Isoform 2 also is a component of dynactin complex from brain, which contains the actin-related protein ARP1.
Heterodimer of an alpha and a beta subunit. Interacts with ARHGAP17 and RCSD1/CAPZIP. Component of the WASH complex, composed of F-actin-capping protein subunit alpha (CAPZA1, CAPZA2 or CAPZA3), F-actin-capping protein subunit beta (CAPZB), WASHC1, WASHC2, WASHC3, WASHC4 and WASHC5. Isoform 2 also is a component of dynactin complex from brain, which contains the actin-related protein ARP1. Interacts with ACTG1. Directly interacts with CRAD/KIAA1211; this interaction decreases binding to actin (By similarity).
Heterodimer of an alpha and a beta subunit. Interacts with ARHGAP17 (PubMed:16678097). Interaction with RCSD1/CAPZIP (PubMed:15850461). Component of the WASH complex, composed of F-actin-capping protein subunit alpha (CAPZA1, CAPZA2 or CAPZA3), F-actin-capping protein subunit beta (CAPZB), WASH (WASHC1, WASH2P, WASH3P, WASH4P, WASH5P or WASH6P), WASHC2 (WASHC2A or WASHC2C), WASHC3, WASHC4 and WASHC5 (PubMed:19922875). Interacts with ACTG1 (PubMed:28493397). Directly interacts with CRAD/KIAA1211; this interaction decreases binding to actin (PubMed:30361697).
Heterodimer of an alpha and a beta subunit. Interacts with ARHGAP17 and RCSD1/CAPZIP. Component of the WASH complex, composed of F-actin-capping protein subunit alpha (CAPZA1, CAPZA2 or CAPZA3), F-actin-capping protein subunit beta (CAPZB), WASHC1, WASHC2, WASHC3, WASHC4 and WASHC5. Interacts with ACTG1. Directly interacts with CRAD/KIAA1211; this interaction decreases binding to actin (By similarity).
Heterodimer of an alpha and a beta subunit. Component of the WASH complex (By similarity). Isoform 2 also is a component of dynactin complex from brain, which contains the actin-related protein ARP1.
Heterodimer of an alpha and a beta subunit. Interacts with ARHGAP17 and RCSD1/CAPZIP. Component of the WASH complex, composed of F-actin-capping protein subunit alpha (CAPZA1, CAPZA2 or CAPZA3), F-actin-capping protein subunit beta (CAPZB), WASHC1, WASHC2, WASHC3, WASHC4 and WASHC5. Isoform 2 also is a component of dynactin complex from brain, which contains the actin-related protein ARP1. Interacts with ACTG1. Directly interacts with CRAD/KIAA1211; this interaction decreases binding to actin (By similarity).
Heterodimer of an alpha and a beta subunit. Interacts with ARHGAP17 (PubMed:16678097). Interaction with RCSD1/CAPZIP (PubMed:15850461). Component of the WASH complex, composed of F-actin-capping protein subunit alpha (CAPZA1, CAPZA2 or CAPZA3), F-actin-capping protein subunit beta (CAPZB), WASH (WASHC1, WASH2P, WASH3P, WASH4P, WASH5P or WASH6P), WASHC2 (WASHC2A or WASHC2C), WASHC3, WASHC4 and WASHC5 (PubMed:19922875). Interacts with ACTG1 (PubMed:28493397). Directly interacts with CRAD/KIAA1211; this interaction decreases binding to actin (PubMed:30361697).
Heterodimer of an alpha and a beta subunit. Interacts with ARHGAP17 and RCSD1/CAPZIP. Component of the WASH complex, composed of F-actin-capping protein subunit alpha (CAPZA1, CAPZA2 or CAPZA3), F-actin-capping protein subunit beta (CAPZB), WASHC1, WASHC2, WASHC3, WASHC4 and WASHC5. Interacts with ACTG1. Directly interacts with CRAD/KIAA1211; this interaction decreases binding to actin (By similarity).
Similarity
Belongs to the F-actin-capping protein beta subunit family.
Keywords
3D-structure
Acetylation
Actin capping
Actin-binding
Alternative splicing
Complete proteome
Cytoplasm
Cytoskeleton
Direct protein sequencing
Reference proteome
Phosphoprotein
Coiled coil
Feature
chain F-actin-capping protein subunit beta
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JIS9
Q1HPW1
A0A1E1WJ21
A0A194R544
A0A194QBF5
A0A2A4JDG3
+ More
A0A212ESG1 A0A1B6E0M4 A0A2P8Y6P0 A0A2J7R613 A0A067RE83 E2AEJ3 K7IUS4 A0A0C9REI9 A0A026W7J4 A0A2A3ERK9 V9IH62 A0A088ALS0 E9IJD6 A0A195FLK4 A0A1S3HLK8 A0A195CQT2 T1ISE5 A0A232F0D8 A0A0M9A506 A0A0P4WB08 A0A1J1IW60 A0A182QPP8 U5ELN2 A0A195EN29 F4WRT9 A0A195BUV7 E2BT35 A0A158NEM1 A0A151XIQ7 A0A3L8DAS2 A0A2P2HX07 A0A182RBT4 A0A1Y1LI08 T1PER4 A0A182SZ68 A0A0K8TTY9 A0A1B6JAJ8 A0A182J5C2 A0A182W9W2 A0A1S3DD83 T1E7W6 A0A2M3Z683 A0A2M4AJV2 W5J6I8 A0A336M726 T1E2V4 A0A1L8E5A9 A0A0L7RHI0 A0A1L8EFX0 A0A341BUW6 A0A2Y9LQW2 A0A1Q3FF96 B0WTE9 K7G5K9 A0A182UQT7 A0A182WTF0 A0A182L299 A0A1S4GX64 P14315-2 A0A1V4KR17 B5G462 A0PFK4 A0A182JWA5 I7GUD2 A0A154PIW7 A0A2Y9DME8 A0A1B0CWU4 P47757-2 P47756-2 P79136-2 A0A1S3GLX3 Q923G3 A0A341BRY2 A0A2U4A086 A0A2Y9ELP0 A0A2Y9GKN2 A0A2J8T2D9 A0A2Y9K8L6 A0A2Y9LG10 A0A2U3ZCI2 M3YXJ4 A9XFX6 F7H329 K7AHH7 A0A384MR50 Q5XI32 Q5R507 D2JYW3 A0A286XEV3 A0A310SMK6 E0VDY6 A0A1A9W1R3 A7RU18 A0A0B8RZE9
A0A212ESG1 A0A1B6E0M4 A0A2P8Y6P0 A0A2J7R613 A0A067RE83 E2AEJ3 K7IUS4 A0A0C9REI9 A0A026W7J4 A0A2A3ERK9 V9IH62 A0A088ALS0 E9IJD6 A0A195FLK4 A0A1S3HLK8 A0A195CQT2 T1ISE5 A0A232F0D8 A0A0M9A506 A0A0P4WB08 A0A1J1IW60 A0A182QPP8 U5ELN2 A0A195EN29 F4WRT9 A0A195BUV7 E2BT35 A0A158NEM1 A0A151XIQ7 A0A3L8DAS2 A0A2P2HX07 A0A182RBT4 A0A1Y1LI08 T1PER4 A0A182SZ68 A0A0K8TTY9 A0A1B6JAJ8 A0A182J5C2 A0A182W9W2 A0A1S3DD83 T1E7W6 A0A2M3Z683 A0A2M4AJV2 W5J6I8 A0A336M726 T1E2V4 A0A1L8E5A9 A0A0L7RHI0 A0A1L8EFX0 A0A341BUW6 A0A2Y9LQW2 A0A1Q3FF96 B0WTE9 K7G5K9 A0A182UQT7 A0A182WTF0 A0A182L299 A0A1S4GX64 P14315-2 A0A1V4KR17 B5G462 A0PFK4 A0A182JWA5 I7GUD2 A0A154PIW7 A0A2Y9DME8 A0A1B0CWU4 P47757-2 P47756-2 P79136-2 A0A1S3GLX3 Q923G3 A0A341BRY2 A0A2U4A086 A0A2Y9ELP0 A0A2Y9GKN2 A0A2J8T2D9 A0A2Y9K8L6 A0A2Y9LG10 A0A2U3ZCI2 M3YXJ4 A9XFX6 F7H329 K7AHH7 A0A384MR50 Q5XI32 Q5R507 D2JYW3 A0A286XEV3 A0A310SMK6 E0VDY6 A0A1A9W1R3 A7RU18 A0A0B8RZE9
Pubmed
19121390
26354079
22118469
29403074
24845553
20798317
+ More
20075255 24508170 21282665 28648823 21719571 21347285 30249741 28004739 25315136 26369729 20920257 23761445 24330624 17381049 20966253 12364791 2745461 7929588 2341404 17018643 22951807 19341723 19468303 17242355 21183079 23806337 7665558 16710414 15489334 12665801 15850461 16678097 19413330 19922875 19608861 21834987 21269460 21406692 22223895 22814378 23186163 24275569 25944712 28493397 30361697 9184090 10349636 11042159 11076861 11217851 12466851 12040188 16141073 17939027 24792893 29420470 30723633 25243066 19343716 22673903 21993624 20566863 17615350 25476704
20075255 24508170 21282665 28648823 21719571 21347285 30249741 28004739 25315136 26369729 20920257 23761445 24330624 17381049 20966253 12364791 2745461 7929588 2341404 17018643 22951807 19341723 19468303 17242355 21183079 23806337 7665558 16710414 15489334 12665801 15850461 16678097 19413330 19922875 19608861 21834987 21269460 21406692 22223895 22814378 23186163 24275569 25944712 28493397 30361697 9184090 10349636 11042159 11076861 11217851 12466851 12040188 16141073 17939027 24792893 29420470 30723633 25243066 19343716 22673903 21993624 20566863 17615350 25476704
EMBL
BABH01041409
DQ443291
ABF51380.1
GDQN01004072
JAT86982.1
KQ460685
+ More
KPJ12802.1 KQ459249 KPJ02320.1 NWSH01001909 PCG69718.1 AGBW02012799 OWR44420.1 GEDC01031115 GEDC01016116 GEDC01015627 GEDC01005871 GEDC01003145 GEDC01002078 GEDC01000542 JAS06183.1 JAS21182.1 JAS21671.1 JAS31427.1 JAS34153.1 JAS35220.1 JAS36756.1 PYGN01000864 PSN39920.1 NEVH01006983 PNF36279.1 KK852561 KDR21348.1 GL438862 EFN68150.1 AAZX01005919 GBYB01006750 JAG76517.1 KK107356 EZA52025.1 KZ288191 PBC34433.1 JR044042 AEY59836.1 GL763764 EFZ19314.1 KQ981490 KYN41216.1 KQ977381 KYN03103.1 JH431430 NNAY01001398 OXU24121.1 KQ435756 KOX75989.1 GDRN01056479 JAI65818.1 CVRI01000058 CRL02777.1 AXCN02000864 GANO01001334 JAB58537.1 KQ978625 KYN29576.1 GL888292 EGI63096.1 KQ976401 KYM92367.1 GL450325 EFN81165.1 ADTU01013498 KQ982080 KYQ60303.1 QOIP01000010 RLU17434.1 IACF01000392 LAB66170.1 GEZM01057027 JAV72498.1 KA647272 AFP61901.1 GDAI01000015 JAI17588.1 GECU01011571 JAS96135.1 GAMD01002692 JAA98898.1 GGFM01003253 MBW24004.1 GGFK01007742 MBW41063.1 ADMH02002133 GGFL01004378 ETN58455.1 MBW68556.1 UFQS01000618 UFQT01000618 SSX05461.1 SSX25820.1 GALA01000950 JAA93902.1 GFDF01000196 JAV13888.1 KQ414592 KOC70186.1 GFDG01001300 JAV17499.1 GFDL01008821 JAV26224.1 DS232085 EDS34387.1 AGCU01032938 AGCU01032939 AGCU01032940 AGCU01032941 AGCU01032942 AGCU01032943 AGCU01032944 AGCU01032945 AGCU01032946 AGCU01032947 AAAB01008964 J04959 U07826 LSYS01002427 OPJ86237.1 DQ217035 DQ217036 ACH46073.1 AM410993 CAL69434.1 AB710463 BAM34023.1 KQ434905 KZC11258.1 AJWK01032871 U10406 U10407 FJ692320 AL807811 U03271 BT019470 BT019471 AL035413 AL359199 AL445163 CH471134 BC024601 BC107752 BC109241 BC109242 Z85980 Y10372 BC102613 BC002053 AK156778 AK168738 CH466615 AAH02053.1 BAE33851.1 BAE40579.1 EDL13298.1 NDHI03003525 PNJ27188.1 AEYP01008404 AEYP01008405 AEYP01008406 AEYP01008407 AEYP01008408 AEYP01008409 AEYP01008410 AEMK02000045 EF202986 EF202989 JX569751 JX966416 DQIR01151511 DQIR01207860 DQIR01250994 DQIR01255040 DQIR01297729 DQIR01312228 ABQ96220.1 AGO58802.1 HDB06988.1 GAMT01005124 GAMS01008249 GAMR01003909 GAMQ01001417 GAMP01006171 JAB06737.1 JAB14887.1 JAB30023.1 JAB40434.1 JAB46584.1 GABF01004689 NBAG03000373 JAA17456.1 PNI33496.1 GQ900984 ADO22501.1 BC083861 CR861078 GU165833 ACZ57956.1 AAKN02028758 AAKN02028759 KQ761155 OAD58132.1 DS235088 EEB11592.1 DS469539 EDO44936.1 GBSH01001031 GBSH01000547 JAG68478.1
KPJ12802.1 KQ459249 KPJ02320.1 NWSH01001909 PCG69718.1 AGBW02012799 OWR44420.1 GEDC01031115 GEDC01016116 GEDC01015627 GEDC01005871 GEDC01003145 GEDC01002078 GEDC01000542 JAS06183.1 JAS21182.1 JAS21671.1 JAS31427.1 JAS34153.1 JAS35220.1 JAS36756.1 PYGN01000864 PSN39920.1 NEVH01006983 PNF36279.1 KK852561 KDR21348.1 GL438862 EFN68150.1 AAZX01005919 GBYB01006750 JAG76517.1 KK107356 EZA52025.1 KZ288191 PBC34433.1 JR044042 AEY59836.1 GL763764 EFZ19314.1 KQ981490 KYN41216.1 KQ977381 KYN03103.1 JH431430 NNAY01001398 OXU24121.1 KQ435756 KOX75989.1 GDRN01056479 JAI65818.1 CVRI01000058 CRL02777.1 AXCN02000864 GANO01001334 JAB58537.1 KQ978625 KYN29576.1 GL888292 EGI63096.1 KQ976401 KYM92367.1 GL450325 EFN81165.1 ADTU01013498 KQ982080 KYQ60303.1 QOIP01000010 RLU17434.1 IACF01000392 LAB66170.1 GEZM01057027 JAV72498.1 KA647272 AFP61901.1 GDAI01000015 JAI17588.1 GECU01011571 JAS96135.1 GAMD01002692 JAA98898.1 GGFM01003253 MBW24004.1 GGFK01007742 MBW41063.1 ADMH02002133 GGFL01004378 ETN58455.1 MBW68556.1 UFQS01000618 UFQT01000618 SSX05461.1 SSX25820.1 GALA01000950 JAA93902.1 GFDF01000196 JAV13888.1 KQ414592 KOC70186.1 GFDG01001300 JAV17499.1 GFDL01008821 JAV26224.1 DS232085 EDS34387.1 AGCU01032938 AGCU01032939 AGCU01032940 AGCU01032941 AGCU01032942 AGCU01032943 AGCU01032944 AGCU01032945 AGCU01032946 AGCU01032947 AAAB01008964 J04959 U07826 LSYS01002427 OPJ86237.1 DQ217035 DQ217036 ACH46073.1 AM410993 CAL69434.1 AB710463 BAM34023.1 KQ434905 KZC11258.1 AJWK01032871 U10406 U10407 FJ692320 AL807811 U03271 BT019470 BT019471 AL035413 AL359199 AL445163 CH471134 BC024601 BC107752 BC109241 BC109242 Z85980 Y10372 BC102613 BC002053 AK156778 AK168738 CH466615 AAH02053.1 BAE33851.1 BAE40579.1 EDL13298.1 NDHI03003525 PNJ27188.1 AEYP01008404 AEYP01008405 AEYP01008406 AEYP01008407 AEYP01008408 AEYP01008409 AEYP01008410 AEMK02000045 EF202986 EF202989 JX569751 JX966416 DQIR01151511 DQIR01207860 DQIR01250994 DQIR01255040 DQIR01297729 DQIR01312228 ABQ96220.1 AGO58802.1 HDB06988.1 GAMT01005124 GAMS01008249 GAMR01003909 GAMQ01001417 GAMP01006171 JAB06737.1 JAB14887.1 JAB30023.1 JAB40434.1 JAB46584.1 GABF01004689 NBAG03000373 JAA17456.1 PNI33496.1 GQ900984 ADO22501.1 BC083861 CR861078 GU165833 ACZ57956.1 AAKN02028758 AAKN02028759 KQ761155 OAD58132.1 DS235088 EEB11592.1 DS469539 EDO44936.1 GBSH01001031 GBSH01000547 JAG68478.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000245037
+ More
UP000235965 UP000027135 UP000000311 UP000002358 UP000053097 UP000242457 UP000005203 UP000078541 UP000085678 UP000078542 UP000215335 UP000053105 UP000183832 UP000075886 UP000078492 UP000007755 UP000078540 UP000008237 UP000005205 UP000075809 UP000279307 UP000075900 UP000095301 UP000075901 UP000075880 UP000075920 UP000079169 UP000000673 UP000053825 UP000252040 UP000248483 UP000002320 UP000007267 UP000075903 UP000076407 UP000075882 UP000000539 UP000190648 UP000075881 UP000076502 UP000248480 UP000092461 UP000000589 UP000005640 UP000009136 UP000081671 UP000245320 UP000248484 UP000248481 UP000248482 UP000245340 UP000000715 UP000008227 UP000008225 UP000002494 UP000001595 UP000005447 UP000009046 UP000091820 UP000001593
UP000235965 UP000027135 UP000000311 UP000002358 UP000053097 UP000242457 UP000005203 UP000078541 UP000085678 UP000078542 UP000215335 UP000053105 UP000183832 UP000075886 UP000078492 UP000007755 UP000078540 UP000008237 UP000005205 UP000075809 UP000279307 UP000075900 UP000095301 UP000075901 UP000075880 UP000075920 UP000079169 UP000000673 UP000053825 UP000252040 UP000248483 UP000002320 UP000007267 UP000075903 UP000076407 UP000075882 UP000000539 UP000190648 UP000075881 UP000076502 UP000248480 UP000092461 UP000000589 UP000005640 UP000009136 UP000081671 UP000245320 UP000248484 UP000248481 UP000248482 UP000245340 UP000000715 UP000008227 UP000008225 UP000002494 UP000001595 UP000005447 UP000009046 UP000091820 UP000001593
Pfam
PF01115 F_actin_cap_B
Interpro
SUPFAM
SSF90096
SSF90096
Gene 3D
ProteinModelPortal
H9JIS9
Q1HPW1
A0A1E1WJ21
A0A194R544
A0A194QBF5
A0A2A4JDG3
+ More
A0A212ESG1 A0A1B6E0M4 A0A2P8Y6P0 A0A2J7R613 A0A067RE83 E2AEJ3 K7IUS4 A0A0C9REI9 A0A026W7J4 A0A2A3ERK9 V9IH62 A0A088ALS0 E9IJD6 A0A195FLK4 A0A1S3HLK8 A0A195CQT2 T1ISE5 A0A232F0D8 A0A0M9A506 A0A0P4WB08 A0A1J1IW60 A0A182QPP8 U5ELN2 A0A195EN29 F4WRT9 A0A195BUV7 E2BT35 A0A158NEM1 A0A151XIQ7 A0A3L8DAS2 A0A2P2HX07 A0A182RBT4 A0A1Y1LI08 T1PER4 A0A182SZ68 A0A0K8TTY9 A0A1B6JAJ8 A0A182J5C2 A0A182W9W2 A0A1S3DD83 T1E7W6 A0A2M3Z683 A0A2M4AJV2 W5J6I8 A0A336M726 T1E2V4 A0A1L8E5A9 A0A0L7RHI0 A0A1L8EFX0 A0A341BUW6 A0A2Y9LQW2 A0A1Q3FF96 B0WTE9 K7G5K9 A0A182UQT7 A0A182WTF0 A0A182L299 A0A1S4GX64 P14315-2 A0A1V4KR17 B5G462 A0PFK4 A0A182JWA5 I7GUD2 A0A154PIW7 A0A2Y9DME8 A0A1B0CWU4 P47757-2 P47756-2 P79136-2 A0A1S3GLX3 Q923G3 A0A341BRY2 A0A2U4A086 A0A2Y9ELP0 A0A2Y9GKN2 A0A2J8T2D9 A0A2Y9K8L6 A0A2Y9LG10 A0A2U3ZCI2 M3YXJ4 A9XFX6 F7H329 K7AHH7 A0A384MR50 Q5XI32 Q5R507 D2JYW3 A0A286XEV3 A0A310SMK6 E0VDY6 A0A1A9W1R3 A7RU18 A0A0B8RZE9
A0A212ESG1 A0A1B6E0M4 A0A2P8Y6P0 A0A2J7R613 A0A067RE83 E2AEJ3 K7IUS4 A0A0C9REI9 A0A026W7J4 A0A2A3ERK9 V9IH62 A0A088ALS0 E9IJD6 A0A195FLK4 A0A1S3HLK8 A0A195CQT2 T1ISE5 A0A232F0D8 A0A0M9A506 A0A0P4WB08 A0A1J1IW60 A0A182QPP8 U5ELN2 A0A195EN29 F4WRT9 A0A195BUV7 E2BT35 A0A158NEM1 A0A151XIQ7 A0A3L8DAS2 A0A2P2HX07 A0A182RBT4 A0A1Y1LI08 T1PER4 A0A182SZ68 A0A0K8TTY9 A0A1B6JAJ8 A0A182J5C2 A0A182W9W2 A0A1S3DD83 T1E7W6 A0A2M3Z683 A0A2M4AJV2 W5J6I8 A0A336M726 T1E2V4 A0A1L8E5A9 A0A0L7RHI0 A0A1L8EFX0 A0A341BUW6 A0A2Y9LQW2 A0A1Q3FF96 B0WTE9 K7G5K9 A0A182UQT7 A0A182WTF0 A0A182L299 A0A1S4GX64 P14315-2 A0A1V4KR17 B5G462 A0PFK4 A0A182JWA5 I7GUD2 A0A154PIW7 A0A2Y9DME8 A0A1B0CWU4 P47757-2 P47756-2 P79136-2 A0A1S3GLX3 Q923G3 A0A341BRY2 A0A2U4A086 A0A2Y9ELP0 A0A2Y9GKN2 A0A2J8T2D9 A0A2Y9K8L6 A0A2Y9LG10 A0A2U3ZCI2 M3YXJ4 A9XFX6 F7H329 K7AHH7 A0A384MR50 Q5XI32 Q5R507 D2JYW3 A0A286XEV3 A0A310SMK6 E0VDY6 A0A1A9W1R3 A7RU18 A0A0B8RZE9
PDB
6F3A
E-value=6.8783e-123,
Score=1126
Ontologies
GO
GO:0003779
GO:0005737
GO:0030036
GO:0008290
GO:0051016
GO:0098686
GO:0030018
GO:0030032
GO:0005829
GO:0098685
GO:0022604
GO:0051015
GO:0016020
GO:0030863
GO:0005903
GO:0010591
GO:0030027
GO:0007010
GO:0014069
GO:0051490
GO:0000902
GO:0031115
GO:0014704
GO:0031175
GO:0043025
GO:0043197
GO:0048747
GO:0032279
GO:0033150
GO:0048487
GO:0090036
GO:0071203
GO:0030030
GO:0006888
GO:0015629
GO:0019886
GO:0070062
GO:0045296
GO:0007596
GO:0005856
GO:0030017
GO:0008270
GO:0005634
GO:0006334
GO:0016021
PANTHER
Topology
Subcellular location
Cytoplasm In cardiac muscle, isoform 2 is located at sarcomere intercalated disks. With evidence from 5 publications.
Myofibril In cardiac muscle, isoform 2 is located at sarcomere intercalated disks. With evidence from 5 publications.
Sarcomere In cardiac muscle, isoform 2 is located at sarcomere intercalated disks. With evidence from 5 publications.
I band In cardiac muscle, isoform 2 is located at sarcomere intercalated disks. With evidence from 5 publications.
Cytoskeleton In cardiac muscle, isoform 2 is located at sarcomere intercalated disks. With evidence from 5 publications.
Z line In cardiac muscle, isoform 1 is located at Z-disks of sarcomeres while isoform 2 is enriched at intercalated disks. With evidence from 2 publications.
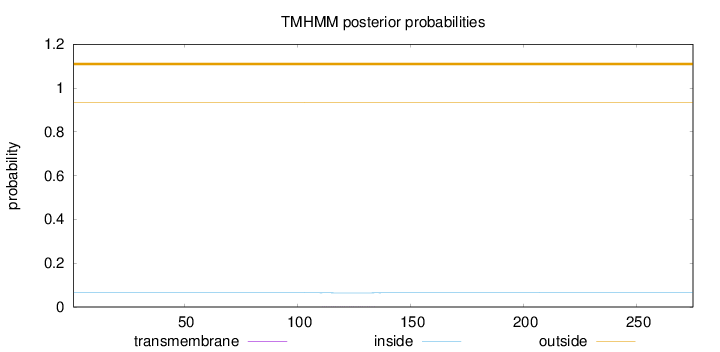
Length:
275
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01302
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06481
outside
1 - 275
Population Genetic Test Statistics
Pi
21.046474
Theta
16.613109
Tajima's D
-1.216345
CLR
2.950722
CSRT
0.0996450177491125
Interpretation
Uncertain
