Gene
KWMTBOMO01124
Annotation
atypical_protein_kinase_C_[Bombyx_mori]
Full name
Protein kinase C
+ More
Atypical protein kinase C
Atypical protein kinase C
Location in the cell
Cytoplasmic Reliability : 2.899
Sequence
CDS
ATGGCACTTGGGACTTTATTGATCGATTGCGAAAGTGAAGACCCGACAAGGGTATTCGGAGTTTATTTGGGGAGCATATACAGGAGAGGAGCACGGCGGTGGCGTAAACTGTACCGCGTCAATGGACACATCTTTCAAGCTAAAAGATTTAATAGACGCGCTTTCTGTGCCTTCTGCCAAGACCGCATCTGGGGCCTCGGAAGACAGGGCTTCAAATGCATCCAGTGTAAGCTGCTCGTTCACAAGAAGTGTCACAAGCTCGTGCAGAAGCCGTGCTCGAATGAGCATGTCGATCCCATTGAGGTCAAAGACGACGCGAACGGAGAGGCCACTTTGGGCCGGGCTTCTTCTGTCAGGTCTCGTGCGGAGGAACAGCCCCCTCTGCCGGAGACGCCGCCGGCGCCGGCGCCCCCCGCCCGCCCCGAGGAGCTGGAGCCGGGCTCGCAGCGGCAGTACTCGCTCGACGACTTTGAGCTCATCAGGGTCATCGGACGTGGCTCTTATGCCAAGGTGCTGATGGTAGAATTAAAACGCACGAAGCGCATATACGCTATGAAGGTGATAAAGAAGGCGCTCGTCACCGACGACGAGGACATCGACTGGGTTCAGACTGAGAAACACGTGTTCGAGACCGCCTCGAACCACCCATTCCTGGTCGGCCTGCACTCCTGCTTCCAAACACCGAGCAGATTGTTCTTCGTGATCGAGTTCGTCAGGGGAGGGGATCTGATGTTCCACATGCAGCGTCAGCGGCGGCTGCCGGAGGAGCACGCGCGGTTCTACGCGGCCGAGATCTCGCTGGCGCTGCACTTCCTGCACGAGCGCGGCGTCATCTACCGCGACCTCAAGCTGGACAACGTGCTGCTCGACCACGAGGGACACATCAAGCTCACCGATTACGGAATGTGCAAGGAGGGAGTGCGTCCGGGCGACACGACCTCTACGTTCTGCGGCACTCCCAACTACATCGCGCCGGAGATCCTCCGCGGCGAGGAGTACGGCTTCTCCGTGGACTGGTGGGCGCTCGGGGTCCTCACGTACGAGATGTTGGCTGGCAGATCGCCCTTCGACATCGCCCAAGCTGCTGACAACCCCGACCAGAATACAGAAGACTATCTCTTCCAGGTGATATTGGAGAAGACGATCCGTATCCCGCGCTCGCTGTCCGTGAAGGCGGCCTCGGTGCTGAAGGGCTTCCTCAACAAGAACCCGGTGGAGCGGCTCGGCTGCGGCGAGAACGGCTTCCTCGACATCGTCAACCATCCCTTCTTCAAGAGCATCGAGTGGGAGATGTTGGAGCAGAAACAAGTGGTGCCTCCGTTCAAGCCCCGTCTGGAAGGCGAACGGGACCTCGCCAACTTCCCGCCGGAGTTCACTGACGAGCCCGTGCATCTCACGCCCGATAACGAGAGCGTGATCGCCGACATCGACCAGTCGGAGTTCGAAGGGTTCGAGTACGTGAACCCGCTGCTGATGTCCCTGGAGGACTGCGTGTGA
Protein
MALGTLLIDCESEDPTRVFGVYLGSIYRRGARRWRKLYRVNGHIFQAKRFNRRAFCAFCQDRIWGLGRQGFKCIQCKLLVHKKCHKLVQKPCSNEHVDPIEVKDDANGEATLGRASSVRSRAEEQPPLPETPPAPAPPARPEELEPGSQRQYSLDDFELIRVIGRGSYAKVLMVELKRTKRIYAMKVIKKALVTDDEDIDWVQTEKHVFETASNHPFLVGLHSCFQTPSRLFFVIEFVRGGDLMFHMQRQRRLPEEHARFYAAEISLALHFLHERGVIYRDLKLDNVLLDHEGHIKLTDYGMCKEGVRPGDTTSTFCGTPNYIAPEILRGEEYGFSVDWWALGVLTYEMLAGRSPFDIAQAADNPDQNTEDYLFQVILEKTIRIPRSLSVKAASVLKGFLNKNPVERLGCGENGFLDIVNHPFFKSIEWEMLEQKQVVPPFKPRLEGERDLANFPPEFTDEPVHLTPDNESVIADIDQSEFEGFEYVNPLLMSLEDCV
Summary
Description
Serine/threonine protein kinase which is required for apico-basal cell polarity in the germ line as well as in epithelial and neural precursor cells, for epithelial planar cell polarity and for cell proliferation. During oocyte development, required for the posterior translocation of oocyte specification factors and for the posterior establishment of the microtubule organizing center within the presumptive oocyte. Phosphorylates l(2)gl which restricts l(2)gl activity to the oocyte posterior and regulates posterior enrichment of par-1, leading to establishment of correct oocyte polarity. Essential for apical localization of l(2)gl and par-6 in neuroblasts and for exclusion of mira from the apical cortex. Phosphorylates baz which is required for targeting of baz to the postsynaptic region where it is involved in actin organization, and for apical exclusion of baz which is necessary for establishment of the apical/lateral border in epithelial cells. Phosphorylates yrt which prevents its premature apical localization and is necessary for correct epithelial cell polarization. Required for the establishment of mitotic spindle orientation during symmetric division of epithelial cells and for apical exclusion of raps/Pins. Involved in symmetric adherens junction positioning during embryogenesis. Required for polarization of the spermatid cyst which is necessary for sperm differentiation. Required for stimulation of the Toll signaling pathway which activates Dif and dl and plays a role in innate immunity. Plays a role in memory enhancement.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Interacts with baz; the interaction is required for apical localization of aPKC in neuroblasts and epithelial cells. Interacts with Dap160; the interaction promotes aPKC apical localization and kinase activity. Interacts with and phosphorylates l(2)gl and yrt. Interacts with crb and ref(2)P. Forms a complex with baz, fz and Patj.
Similarity
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. PKC subfamily.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family.
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family.
Belongs to the protein kinase superfamily.
Keywords
Alternative splicing
ATP-binding
Cell membrane
Complete proteome
Cytoplasm
Kinase
Membrane
Metal-binding
Nucleotide-binding
Reference proteome
Transferase
Zinc
Zinc-finger
Feature
chain Protein kinase C
splice variant In isoform F.
splice variant In isoform F.
Uniprot
A0A2R8G5Z7
Q4AED5
A0A2A4JY12
A0A2A4JZT3
A0A1L8DMH1
A0A1L8DMD5
+ More
A0A1Q3G2J3 A0A1Q3G2D1 A0A067R2P7 A0A1W4XKZ1 A0A1B6DB11 A0A1W4XL00 A0A023F5R6 A0A224X9U0 A0A182QX87 Q17GR4 A0A1W4XJV6 A0A182IQT2 A0A0P6IYY5 A0A2M4CTW5 A0A2M4AHK3 A0A2M3Z5G1 A0A1B6KEU6 A0A1B6KX77 A0A1Y1MDI0 A0A1Y1MBV0 A0A0A9ZEF0 A0A0A9ZC39 V5GWH4 A0A034VKR0 A0A1I8M1P2 V5GSK4 A0A0K8WKF8 A0A0M3TCH4 A0A1A9Y727 A0A1A9Z5B8 B4KP34 A0A1A9WV58 A0A310S6P5 A0A0J9RD44 A0A088A8P9 A0A0J9RCU8 A0A0L7REQ1 A0A0B4JD78 A0A0B4KG15 A0A0Q9WFW4 B4HRS5 V9I8D2 A0A0Q5W036 A0A0Q5VLS2 A0A0J9RD09 A0A0B4KEQ8 A0A0B4KEV2 A0A0R1DRI0 A0A1I8PAU2 V9IAN8 A1Z9X0 A0A2A3EGP3 A0A154P8H9 A0A2S2PS44 A0A1W4U9P8 A0A1W4ULK7 A1Z9X0-4 A0A0J9RCV4 N6WCA0 A0A0B4KFQ6 A0A0P8ZLZ9 B4J5Z1 A1Z9X0-5 A0A182UEM5 A0A1W4U9Q2 Q28YC5 A0A3B0J2S6 B3MH03 A0A1W4ULS4 X1WYI0 N6VFF2 A0A1W4UAB5 A0A2S2QHV1 K7J023 A0A182KLK6 A0A182FVS2 B4MQ00 E0VZB9 A0A194R4Q6 A0A158NZJ5 A0A182HHR7 A0A195FDA6 A0A182M485 A0A195EK64 A0A2J7R615 E2BWJ8 A0A0P4W3P1 E9GL76 A0A1S4H907 T1J7C8 A0A2H4IAU8 A0A0K2TS93 A0A0J9RD05 A0A3L8D787
A0A1Q3G2J3 A0A1Q3G2D1 A0A067R2P7 A0A1W4XKZ1 A0A1B6DB11 A0A1W4XL00 A0A023F5R6 A0A224X9U0 A0A182QX87 Q17GR4 A0A1W4XJV6 A0A182IQT2 A0A0P6IYY5 A0A2M4CTW5 A0A2M4AHK3 A0A2M3Z5G1 A0A1B6KEU6 A0A1B6KX77 A0A1Y1MDI0 A0A1Y1MBV0 A0A0A9ZEF0 A0A0A9ZC39 V5GWH4 A0A034VKR0 A0A1I8M1P2 V5GSK4 A0A0K8WKF8 A0A0M3TCH4 A0A1A9Y727 A0A1A9Z5B8 B4KP34 A0A1A9WV58 A0A310S6P5 A0A0J9RD44 A0A088A8P9 A0A0J9RCU8 A0A0L7REQ1 A0A0B4JD78 A0A0B4KG15 A0A0Q9WFW4 B4HRS5 V9I8D2 A0A0Q5W036 A0A0Q5VLS2 A0A0J9RD09 A0A0B4KEQ8 A0A0B4KEV2 A0A0R1DRI0 A0A1I8PAU2 V9IAN8 A1Z9X0 A0A2A3EGP3 A0A154P8H9 A0A2S2PS44 A0A1W4U9P8 A0A1W4ULK7 A1Z9X0-4 A0A0J9RCV4 N6WCA0 A0A0B4KFQ6 A0A0P8ZLZ9 B4J5Z1 A1Z9X0-5 A0A182UEM5 A0A1W4U9Q2 Q28YC5 A0A3B0J2S6 B3MH03 A0A1W4ULS4 X1WYI0 N6VFF2 A0A1W4UAB5 A0A2S2QHV1 K7J023 A0A182KLK6 A0A182FVS2 B4MQ00 E0VZB9 A0A194R4Q6 A0A158NZJ5 A0A182HHR7 A0A195FDA6 A0A182M485 A0A195EK64 A0A2J7R615 E2BWJ8 A0A0P4W3P1 E9GL76 A0A1S4H907 T1J7C8 A0A2H4IAU8 A0A0K2TS93 A0A0J9RD05 A0A3L8D787
EC Number
2.7.11.13
Pubmed
24845553
25474469
17510324
26999592
28004739
25401762
+ More
26823975 25348373 25315136 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 10995441 12537569 11734648 12446795 11914720 14657233 15302858 15907474 17488624 18094021 18614576 19472188 20434988 22223679 24515345 24830287 15632085 18057021 20075255 20966253 20566863 26354079 21347285 20798317 21292972 30249741
26823975 25348373 25315136 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 10995441 12537569 11734648 12446795 11914720 14657233 15302858 15907474 17488624 18094021 18614576 19472188 20434988 22223679 24515345 24830287 15632085 18057021 20075255 20966253 20566863 26354079 21347285 20798317 21292972 30249741
EMBL
MF040614
AVZ23636.1
AB184962
BAE17023.1
NWSH01000370
PCG76911.1
+ More
PCG76912.1 GFDF01006473 JAV07611.1 GFDF01006472 JAV07612.1 GFDL01001086 JAV33959.1 GFDL01001085 JAV33960.1 KK853053 KDR12017.1 GEDC01014425 GEDC01003682 JAS22873.1 JAS33616.1 GBBI01002193 JAC16519.1 GFTR01007299 JAW09127.1 AXCN02001803 CH477256 EAT45873.1 GDUN01000867 JAN95052.1 GGFL01004614 MBW68792.1 GGFK01006881 MBW40202.1 GGFM01003001 MBW23752.1 GEBQ01029985 GEBQ01025291 JAT09992.1 JAT14686.1 GEBQ01023924 GEBQ01010216 JAT16053.1 JAT29761.1 GEZM01035984 JAV83058.1 GEZM01035982 JAV83061.1 GBHO01000783 GBHO01000780 JAG42821.1 JAG42824.1 GBHO01005115 GBHO01000777 GBHO01000775 GDHC01017225 GDHC01002558 JAG38489.1 JAG42827.1 JAG42829.1 JAQ01404.1 JAQ16071.1 GALX01003878 JAB64588.1 GAKP01016547 GAKP01016545 GAKP01016544 JAC42405.1 GALX01003879 JAB64587.1 GDHF01023493 GDHF01000974 JAI28821.1 JAI51340.1 KR075828 ALE20547.1 CH933808 EDW10100.2 KQ767320 OAD53404.1 CM002911 KMY93886.1 KMY93883.1 KQ414609 KOC69318.1 AE013599 ADV37185.1 AGB93516.1 CH940648 KRF79653.1 CH480816 EDW47939.1 JR036779 AEY57393.1 CH954179 KQS62586.1 KQS62589.1 KMY93887.1 AGB93519.1 AGB93518.1 CM000158 KRJ99614.1 JR036780 AEY57394.1 AF288482 AY118402 BT056247 AAF58177.2 AAG01528.1 AAM48431.1 ACL68694.1 KZ288254 PBC30897.1 KQ434844 KZC08172.1 GGMR01019673 MBY32292.1 KMY93888.1 CM000071 ENO01779.1 AGB93517.1 CH902619 KPU75811.1 CH916367 EDW00834.1 EAL26040.1 OUUW01000001 SPP75537.1 EDV35762.1 KPU75812.1 ABLF02028965 ABLF02028966 ENO01780.2 GGMS01007887 MBY77090.1 CH963849 EDW74189.1 DS235851 EEB18725.1 KQ460685 KPJ12798.1 ADTU01004650 APCN01002436 KQ981685 KYN38024.1 AXCM01002439 KQ978747 KYN28655.1 NEVH01006983 PNF36270.1 GL451131 EFN79954.1 GDRN01074945 JAI63151.1 GL732550 EFX79842.1 JH431917 KX100579 ARV85772.1 HACA01011543 CDW28904.1 KMY93882.1 QOIP01000012 RLU16106.1
PCG76912.1 GFDF01006473 JAV07611.1 GFDF01006472 JAV07612.1 GFDL01001086 JAV33959.1 GFDL01001085 JAV33960.1 KK853053 KDR12017.1 GEDC01014425 GEDC01003682 JAS22873.1 JAS33616.1 GBBI01002193 JAC16519.1 GFTR01007299 JAW09127.1 AXCN02001803 CH477256 EAT45873.1 GDUN01000867 JAN95052.1 GGFL01004614 MBW68792.1 GGFK01006881 MBW40202.1 GGFM01003001 MBW23752.1 GEBQ01029985 GEBQ01025291 JAT09992.1 JAT14686.1 GEBQ01023924 GEBQ01010216 JAT16053.1 JAT29761.1 GEZM01035984 JAV83058.1 GEZM01035982 JAV83061.1 GBHO01000783 GBHO01000780 JAG42821.1 JAG42824.1 GBHO01005115 GBHO01000777 GBHO01000775 GDHC01017225 GDHC01002558 JAG38489.1 JAG42827.1 JAG42829.1 JAQ01404.1 JAQ16071.1 GALX01003878 JAB64588.1 GAKP01016547 GAKP01016545 GAKP01016544 JAC42405.1 GALX01003879 JAB64587.1 GDHF01023493 GDHF01000974 JAI28821.1 JAI51340.1 KR075828 ALE20547.1 CH933808 EDW10100.2 KQ767320 OAD53404.1 CM002911 KMY93886.1 KMY93883.1 KQ414609 KOC69318.1 AE013599 ADV37185.1 AGB93516.1 CH940648 KRF79653.1 CH480816 EDW47939.1 JR036779 AEY57393.1 CH954179 KQS62586.1 KQS62589.1 KMY93887.1 AGB93519.1 AGB93518.1 CM000158 KRJ99614.1 JR036780 AEY57394.1 AF288482 AY118402 BT056247 AAF58177.2 AAG01528.1 AAM48431.1 ACL68694.1 KZ288254 PBC30897.1 KQ434844 KZC08172.1 GGMR01019673 MBY32292.1 KMY93888.1 CM000071 ENO01779.1 AGB93517.1 CH902619 KPU75811.1 CH916367 EDW00834.1 EAL26040.1 OUUW01000001 SPP75537.1 EDV35762.1 KPU75812.1 ABLF02028965 ABLF02028966 ENO01780.2 GGMS01007887 MBY77090.1 CH963849 EDW74189.1 DS235851 EEB18725.1 KQ460685 KPJ12798.1 ADTU01004650 APCN01002436 KQ981685 KYN38024.1 AXCM01002439 KQ978747 KYN28655.1 NEVH01006983 PNF36270.1 GL451131 EFN79954.1 GDRN01074945 JAI63151.1 GL732550 EFX79842.1 JH431917 KX100579 ARV85772.1 HACA01011543 CDW28904.1 KMY93882.1 QOIP01000012 RLU16106.1
Proteomes
UP000218220
UP000027135
UP000192223
UP000075886
UP000008820
UP000075880
+ More
UP000095301 UP000092443 UP000092445 UP000009192 UP000091820 UP000005203 UP000053825 UP000000803 UP000008792 UP000001292 UP000008711 UP000002282 UP000095300 UP000242457 UP000076502 UP000192221 UP000001819 UP000007801 UP000001070 UP000075902 UP000268350 UP000007819 UP000002358 UP000075882 UP000069272 UP000007798 UP000009046 UP000053240 UP000005205 UP000075840 UP000078541 UP000075883 UP000078492 UP000235965 UP000008237 UP000000305 UP000279307
UP000095301 UP000092443 UP000092445 UP000009192 UP000091820 UP000005203 UP000053825 UP000000803 UP000008792 UP000001292 UP000008711 UP000002282 UP000095300 UP000242457 UP000076502 UP000192221 UP000001819 UP000007801 UP000001070 UP000075902 UP000268350 UP000007819 UP000002358 UP000075882 UP000069272 UP000007798 UP000009046 UP000053240 UP000005205 UP000075840 UP000078541 UP000075883 UP000078492 UP000235965 UP000008237 UP000000305 UP000279307
Pfam
Interpro
IPR000961
AGC-kinase_C
+ More
IPR020454 DAG/PE-bd
IPR017892 Pkinase_C
IPR000719 Prot_kinase_dom
IPR034659 Atypical_PKC
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR002219 PE/DAG-bd
IPR034877 PB1_aPKC
IPR012233 PKC
IPR000270 PB1_dom
IPR007110 Ig-like_dom
IPR036179 Ig-like_dom_sf
IPR013783 Ig-like_fold
IPR005829 Sugar_transporter_CS
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035979 RBD_domain_sf
IPR014375 Protein_kinase_C_a/b/g
IPR020454 DAG/PE-bd
IPR017892 Pkinase_C
IPR000719 Prot_kinase_dom
IPR034659 Atypical_PKC
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR002219 PE/DAG-bd
IPR034877 PB1_aPKC
IPR012233 PKC
IPR000270 PB1_dom
IPR007110 Ig-like_dom
IPR036179 Ig-like_dom_sf
IPR013783 Ig-like_fold
IPR005829 Sugar_transporter_CS
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035979 RBD_domain_sf
IPR014375 Protein_kinase_C_a/b/g
Gene 3D
ProteinModelPortal
A0A2R8G5Z7
Q4AED5
A0A2A4JY12
A0A2A4JZT3
A0A1L8DMH1
A0A1L8DMD5
+ More
A0A1Q3G2J3 A0A1Q3G2D1 A0A067R2P7 A0A1W4XKZ1 A0A1B6DB11 A0A1W4XL00 A0A023F5R6 A0A224X9U0 A0A182QX87 Q17GR4 A0A1W4XJV6 A0A182IQT2 A0A0P6IYY5 A0A2M4CTW5 A0A2M4AHK3 A0A2M3Z5G1 A0A1B6KEU6 A0A1B6KX77 A0A1Y1MDI0 A0A1Y1MBV0 A0A0A9ZEF0 A0A0A9ZC39 V5GWH4 A0A034VKR0 A0A1I8M1P2 V5GSK4 A0A0K8WKF8 A0A0M3TCH4 A0A1A9Y727 A0A1A9Z5B8 B4KP34 A0A1A9WV58 A0A310S6P5 A0A0J9RD44 A0A088A8P9 A0A0J9RCU8 A0A0L7REQ1 A0A0B4JD78 A0A0B4KG15 A0A0Q9WFW4 B4HRS5 V9I8D2 A0A0Q5W036 A0A0Q5VLS2 A0A0J9RD09 A0A0B4KEQ8 A0A0B4KEV2 A0A0R1DRI0 A0A1I8PAU2 V9IAN8 A1Z9X0 A0A2A3EGP3 A0A154P8H9 A0A2S2PS44 A0A1W4U9P8 A0A1W4ULK7 A1Z9X0-4 A0A0J9RCV4 N6WCA0 A0A0B4KFQ6 A0A0P8ZLZ9 B4J5Z1 A1Z9X0-5 A0A182UEM5 A0A1W4U9Q2 Q28YC5 A0A3B0J2S6 B3MH03 A0A1W4ULS4 X1WYI0 N6VFF2 A0A1W4UAB5 A0A2S2QHV1 K7J023 A0A182KLK6 A0A182FVS2 B4MQ00 E0VZB9 A0A194R4Q6 A0A158NZJ5 A0A182HHR7 A0A195FDA6 A0A182M485 A0A195EK64 A0A2J7R615 E2BWJ8 A0A0P4W3P1 E9GL76 A0A1S4H907 T1J7C8 A0A2H4IAU8 A0A0K2TS93 A0A0J9RD05 A0A3L8D787
A0A1Q3G2J3 A0A1Q3G2D1 A0A067R2P7 A0A1W4XKZ1 A0A1B6DB11 A0A1W4XL00 A0A023F5R6 A0A224X9U0 A0A182QX87 Q17GR4 A0A1W4XJV6 A0A182IQT2 A0A0P6IYY5 A0A2M4CTW5 A0A2M4AHK3 A0A2M3Z5G1 A0A1B6KEU6 A0A1B6KX77 A0A1Y1MDI0 A0A1Y1MBV0 A0A0A9ZEF0 A0A0A9ZC39 V5GWH4 A0A034VKR0 A0A1I8M1P2 V5GSK4 A0A0K8WKF8 A0A0M3TCH4 A0A1A9Y727 A0A1A9Z5B8 B4KP34 A0A1A9WV58 A0A310S6P5 A0A0J9RD44 A0A088A8P9 A0A0J9RCU8 A0A0L7REQ1 A0A0B4JD78 A0A0B4KG15 A0A0Q9WFW4 B4HRS5 V9I8D2 A0A0Q5W036 A0A0Q5VLS2 A0A0J9RD09 A0A0B4KEQ8 A0A0B4KEV2 A0A0R1DRI0 A0A1I8PAU2 V9IAN8 A1Z9X0 A0A2A3EGP3 A0A154P8H9 A0A2S2PS44 A0A1W4U9P8 A0A1W4ULK7 A1Z9X0-4 A0A0J9RCV4 N6WCA0 A0A0B4KFQ6 A0A0P8ZLZ9 B4J5Z1 A1Z9X0-5 A0A182UEM5 A0A1W4U9Q2 Q28YC5 A0A3B0J2S6 B3MH03 A0A1W4ULS4 X1WYI0 N6VFF2 A0A1W4UAB5 A0A2S2QHV1 K7J023 A0A182KLK6 A0A182FVS2 B4MQ00 E0VZB9 A0A194R4Q6 A0A158NZJ5 A0A182HHR7 A0A195FDA6 A0A182M485 A0A195EK64 A0A2J7R615 E2BWJ8 A0A0P4W3P1 E9GL76 A0A1S4H907 T1J7C8 A0A2H4IAU8 A0A0K2TS93 A0A0J9RD05 A0A3L8D787
PDB
5LIH
E-value=1.27215e-173,
Score=1566
Ontologies
GO
GO:0005524
GO:0046872
GO:0007163
GO:0004697
GO:0035556
GO:0004674
GO:0022857
GO:0016021
GO:1904580
GO:0016324
GO:0030011
GO:0007613
GO:0045179
GO:0000132
GO:0045167
GO:0003382
GO:0045198
GO:0016332
GO:0051601
GO:0005938
GO:0001738
GO:0007314
GO:0002052
GO:0060446
GO:0017022
GO:0055059
GO:0030860
GO:0006468
GO:0035003
GO:0016334
GO:0035011
GO:0034332
GO:0010592
GO:0007423
GO:0045196
GO:0007416
GO:0016327
GO:0007294
GO:0045197
GO:0007043
GO:0045176
GO:0072659
GO:0045186
GO:0046667
GO:0007430
GO:0090163
GO:0007283
GO:0051491
GO:0003723
GO:0008270
GO:0018105
GO:0004672
GO:0005634
GO:0006334
GO:0003779
Topology
Subcellular location
Cytoplasm
Cytoplasmic at interphase but localizes to the apical cell cortex during mitosis. With evidence from 6 publications.
Cell cortex Cytoplasmic at interphase but localizes to the apical cell cortex during mitosis. With evidence from 6 publications.
Apicolateral cell membrane Cytoplasmic at interphase but localizes to the apical cell cortex during mitosis. With evidence from 6 publications.
Cell cortex Cytoplasmic at interphase but localizes to the apical cell cortex during mitosis. With evidence from 6 publications.
Apicolateral cell membrane Cytoplasmic at interphase but localizes to the apical cell cortex during mitosis. With evidence from 6 publications.
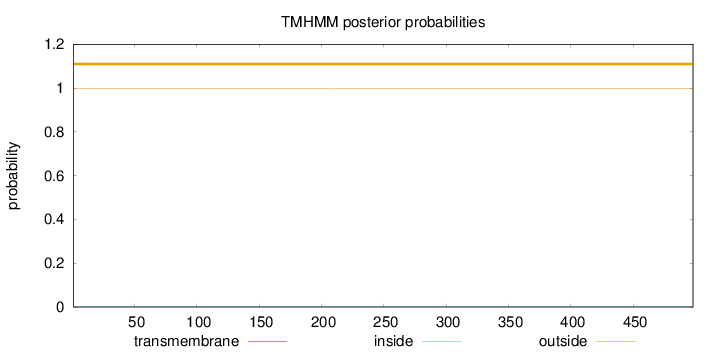
Length:
498
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00348
Exp number, first 60 AAs:
0.00051
Total prob of N-in:
0.00059
outside
1 - 498
Population Genetic Test Statistics
Pi
164.737163
Theta
145.419239
Tajima's D
1.052443
CLR
0.722159
CSRT
0.681565921703915
Interpretation
Uncertain
