Gene
KWMTBOMO01123 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007441
Annotation
PREDICTED:_inositol-3-phosphate_synthase_[Amyelois_transitella]
Full name
Inositol-3-phosphate synthase
Alternative Name
Myo-inositol 1-phosphate synthase
Location in the cell
Cytoplasmic Reliability : 1.907
Sequence
CDS
ATGGAAAAAACACCAAATTTGGTTATAGTCAGTCCGAATGTGAAATATTCGGATGATTTTATTTACGCCGATTACGAGTATAACGAAACATTAGTGAAAAAAATCGACAATCAGTTAGTGGCACAGCCGTACAGCAAAACTTTGAGCATTAGGACTCAGCGTAAAGTTGGCAAAGTTGGAGTGATGCTAGTGGGATGGGGTGGCAACAATGGTTCAACATTTACTGCGGCTGTTTTGGCCAACCGACACCAGTTGTCCTGGAACACAAAGAATGGCTCTCTAGATGCTAATTGGTTTGGTTCGATCACACAAGCATCGACTGTGAGACTGGGGTTAGATGAAAAAGGGAATGATGTGAATGTGCCAATGAACAGTTTACTGCCGATGGTGAACCCTGATGATCTATTTATTGATGGCTGGGACATAAGTCCTCTCAATTTGGCGGAGTCGATGGCTCGAGCTAAGGTCCTCGATTATGATCTGCAGCAGAAACTGAAAAAGGAGATGGCCACTATGAAACCGAGACCAGCTATTTATGATCCTGACTTCATTGCTGCGAATCAGGCCGATCGTGCTACGAACCTCATCCACGGCACGAAGTACGAGCAGTACCTGCAAATCCGCGCCGATATCAAGGACTTCAAGGATAAGAACAAACTGGACAAAGTCATAGTTGTCTGGACGGCGAACACTGAAAGGTTCTGCGAGGTCCGCGACGGCGTGCACGACACGAGCGACAACTTGGAGAAGGCGCTGCGGGACAACGAGCCGGAGATATCGCCGTCCACCATCATCGCGCTCGCTTCGATCGACGAGGGGTGTTGCTATATCAACGGCGCGCCTCAAAACACTTTGATCCCGGGCGTCATAGAGCGGGCCGAGAAACGAGGGGTCTTCGTGGCTGGCGACGACTTCAAATCAGGACAGACCAAGCTTAAGTCAGTACTCGTCGACTTCTTGGTTTCGGCTGGTCTGAAACCGGTGTCGATAGTGAGCTACAACCATCTCGGCAACAACGACGGTAAGAATCTCTCCGCTCCGAAGCAGTTCCGCTCGAAAGAGATCAGCAAGAGCAGCGTGGTGGACGACGCGGTGCAGTCGAACGGCGTGCTGTACCGGCCCGGGGAGCGCCCCGACCACGTGGTCGTCATCAAGTACGTGCCCTACGTCGGTGACTCGAAGCGCGCCATGGACGAGTACACGTCGGAGATCCTGCTGCACGGCACGAACACGCTGGTGGTGCACAACACGTGCGAGGACTCGCTGCTGGCGGCGCCGCTGCTGCTGGACCTGGCGGTGCTGGCGGAGCTGCTGGGGCGCGTGGCGCTGCGGGCGGGCGGCGCGTGGGCGGCGCTGCACCCCGCGCTGTCGCCGCTCGCCTACCTGCTGAAGGCGCCGCTGGTGCCGGCGGGCGCGCCCGTCGTCAACGCGCTGTCCAAGCAGCGCGCCTGCATCGACAACCTGCTGCGCGCCTGCCTCGGCCTGCCCTGTCTACACAACATGCAGCTCGAGCATAAGGTACCGTTTCTAATGAGCGAGCTCCGTGACGGGTCGATGTTCGCCGGCGCCCCTGCGTTTAAGAAGCCCAAATTGGATCAAGAAAATGGCGATCAATAA
Protein
MEKTPNLVIVSPNVKYSDDFIYADYEYNETLVKKIDNQLVAQPYSKTLSIRTQRKVGKVGVMLVGWGGNNGSTFTAAVLANRHQLSWNTKNGSLDANWFGSITQASTVRLGLDEKGNDVNVPMNSLLPMVNPDDLFIDGWDISPLNLAESMARAKVLDYDLQQKLKKEMATMKPRPAIYDPDFIAANQADRATNLIHGTKYEQYLQIRADIKDFKDKNKLDKVIVVWTANTERFCEVRDGVHDTSDNLEKALRDNEPEISPSTIIALASIDEGCCYINGAPQNTLIPGVIERAEKRGVFVAGDDFKSGQTKLKSVLVDFLVSAGLKPVSIVSYNHLGNNDGKNLSAPKQFRSKEISKSSVVDDAVQSNGVLYRPGERPDHVVVIKYVPYVGDSKRAMDEYTSEILLHGTNTLVVHNTCEDSLLAAPLLLDLAVLAELLGRVALRAGGAWAALHPALSPLAYLLKAPLVPAGAPVVNALSKQRACIDNLLRACLGLPCLHNMQLEHKVPFLMSELRDGSMFAGAPAFKKPKLDQENGDQ
Summary
Catalytic Activity
D-glucose 6-phosphate = 1D-myo-inositol 3-phosphate
Cofactor
NAD(+)
Similarity
Belongs to the myo-inositol 1-phosphate synthase family.
Keywords
Complete proteome
Cytoplasm
Inositol biosynthesis
Isomerase
Lipid biosynthesis
Lipid metabolism
NAD
Phospholipid biosynthesis
Phospholipid metabolism
Phosphoprotein
Reference proteome
Feature
chain Inositol-3-phosphate synthase
Uniprot
H9JD44
A0A194QBG0
A0A194R539
A0A2A4JZS2
W8AXX6
A0A2J7RJU5
+ More
A0A1A9WR46 A0A1W4X2G4 A0A034WPS6 A0A0K8VF34 D6WV33 A0A1A9XFG4 A0A1I8PC50 A0A1B0G1H8 A0A1B0CKN3 A0A1A9V3A3 A0A1B0AGG2 A0A067R717 B4J6P6 A0A182GQX6 B4KNC9 A0A1L8DYZ6 Q17H06 A0A182GVS5 A0A0L0BW05 A0A0M4EIF0 T1PP12 B4MR64 A0A1I8NDV0 A0A1B0CZ62 A0A3B0JJW5 A0A182JXK2 Q290L0 B4GBP1 A0A182N3W5 A0A182FUZ8 V5GMK8 A0A1Y1MZR4 A0A182QAP8 B3MDQ6 A0A182HHE3 B4P1P6 A0A182YLA1 A0A182VF33 A0A1J1I609 A0A182XGS6 A0A1S4EBW8 B3N9Z1 A0A336M0U0 A0A1W4V0U6 A0A182MDN0 A0A182RYY1 R7UC15 Q7PZB9 A0A0J9R749 A0A088AHY9 A0A182L4Y4 O97477 A0A1Q3FPL3 V9I9S1 A0A182UEU2 V4B2T7 A0A1S3H4Z5 A0A182P7I1 A0A182W1X7 B0WI11 A0A1Q3FPM1 A0A084WIU1 W5JMX9 B4LMP9 A0A2R5LH56 S7N5Q4 A0A1B6MTK9 A0A087TUU5 A0A182J2U9 A0A1S3IQ99 G9K677 A0A2A3E1Y2 A0A154PE42 U5EV30 A0A158NYD0 M3YVS2 A0A0L7QZJ6 F4WSZ3 E2A4B2 A0A3P9KY71 A0A1X7V072 H2MGD4 A0A195F761 G1NX43 A0A195AYD6 A0A0M8ZNV0 A0A3P9HWT8 L5L7E3 A0A3M6U3B9 E2BZ44 A0A2Y9IYD6 M3WC06 A0A0P6ADI8 A0A1B6JAG6
A0A1A9WR46 A0A1W4X2G4 A0A034WPS6 A0A0K8VF34 D6WV33 A0A1A9XFG4 A0A1I8PC50 A0A1B0G1H8 A0A1B0CKN3 A0A1A9V3A3 A0A1B0AGG2 A0A067R717 B4J6P6 A0A182GQX6 B4KNC9 A0A1L8DYZ6 Q17H06 A0A182GVS5 A0A0L0BW05 A0A0M4EIF0 T1PP12 B4MR64 A0A1I8NDV0 A0A1B0CZ62 A0A3B0JJW5 A0A182JXK2 Q290L0 B4GBP1 A0A182N3W5 A0A182FUZ8 V5GMK8 A0A1Y1MZR4 A0A182QAP8 B3MDQ6 A0A182HHE3 B4P1P6 A0A182YLA1 A0A182VF33 A0A1J1I609 A0A182XGS6 A0A1S4EBW8 B3N9Z1 A0A336M0U0 A0A1W4V0U6 A0A182MDN0 A0A182RYY1 R7UC15 Q7PZB9 A0A0J9R749 A0A088AHY9 A0A182L4Y4 O97477 A0A1Q3FPL3 V9I9S1 A0A182UEU2 V4B2T7 A0A1S3H4Z5 A0A182P7I1 A0A182W1X7 B0WI11 A0A1Q3FPM1 A0A084WIU1 W5JMX9 B4LMP9 A0A2R5LH56 S7N5Q4 A0A1B6MTK9 A0A087TUU5 A0A182J2U9 A0A1S3IQ99 G9K677 A0A2A3E1Y2 A0A154PE42 U5EV30 A0A158NYD0 M3YVS2 A0A0L7QZJ6 F4WSZ3 E2A4B2 A0A3P9KY71 A0A1X7V072 H2MGD4 A0A195F761 G1NX43 A0A195AYD6 A0A0M8ZNV0 A0A3P9HWT8 L5L7E3 A0A3M6U3B9 E2BZ44 A0A2Y9IYD6 M3WC06 A0A0P6ADI8 A0A1B6JAG6
EC Number
5.5.1.4
Pubmed
19121390
26354079
24495485
25348373
18362917
19820115
+ More
24845553 17994087 26483478 17510324 26108605 25315136 15632085 28004739 17550304 25244985 23254933 12364791 22936249 20966253 11121586 10731132 12537572 12537569 17372656 24438588 20920257 23761445 23236062 21347285 21719571 20798317 17554307 21993624 23258410 30382153 17975172
24845553 17994087 26483478 17510324 26108605 25315136 15632085 28004739 17550304 25244985 23254933 12364791 22936249 20966253 11121586 10731132 12537572 12537569 17372656 24438588 20920257 23761445 23236062 21347285 21719571 20798317 17554307 21993624 23258410 30382153 17975172
EMBL
BABH01027813
BABH01027814
BABH01027815
BABH01027816
BABH01027817
KQ459249
+ More
KPJ02325.1 KQ460685 KPJ12797.1 NWSH01000370 PCG76910.1 GAMC01015618 GAMC01015617 JAB90937.1 NEVH01002987 PNF41107.1 GAKP01002615 JAC56337.1 GDHF01014841 JAI37473.1 KQ971357 EFA09280.1 CCAG010005392 AJWK01016483 AJWK01016484 KK852699 KDR18213.1 CH916367 EDW00949.1 JXUM01081571 JXUM01081572 JXUM01081573 KQ563255 KXJ74195.1 CH933808 EDW09982.1 GFDF01002428 JAV11656.1 CH477254 EAT45917.1 EAT45918.1 JXUM01091697 JXUM01091698 JXUM01091699 KQ563935 KXJ73050.1 JRES01001254 KNC24206.1 CP012524 ALC40855.1 KA647170 KA649733 KA649753 AFP64362.1 CH963849 EDW74603.1 AJVK01009514 AJVK01009515 AJVK01009516 OUUW01000001 SPP75670.1 CM000071 EAL25351.1 CH479181 EDW32299.1 GALX01005629 JAB62837.1 GEZM01016729 JAV91162.1 AXCN02001095 CH902619 EDV36441.1 CM000157 EDW89182.1 CVRI01000040 CRK95020.1 CH954177 EDV59687.1 UFQT01000092 SSX19588.1 AXCM01000489 AMQN01008410 KB303059 ELU03519.1 AAAB01008986 EAA00329.3 CM002911 KMY91977.1 AF071103 AF071104 AE013599 AY122137 BT001560 BT001772 GFDL01005494 JAV29551.1 JR037601 AEY57868.1 KB200665 ESP00812.1 DS231941 EDS28093.1 GFDL01005495 JAV29550.1 ATLV01023946 KE525347 KFB50135.1 ADMH02000915 ETN64673.1 CH940648 EDW60990.1 GGLE01004683 MBY08809.1 KE163515 EPQ12449.1 GEBQ01000716 JAT39261.1 KK116827 KFM68884.1 JP011804 AES00402.1 KZ288478 PBC25266.1 KQ434870 KZC09470.1 GANO01002053 JAB57818.1 ADTU01003756 AEYP01051416 KQ414681 KOC63962.1 GL888331 EGI62697.1 GL436621 EFN71722.1 KQ981768 KYN36022.1 AAPE02040677 KQ976701 KYM77211.1 KQ435955 KOX68055.1 KB030270 ELK19191.1 RCHS01002306 RMX48175.1 GL451555 EFN79046.1 AANG04000016 GDIP01044160 JAM59555.1 GECU01011475 JAS96231.1
KPJ02325.1 KQ460685 KPJ12797.1 NWSH01000370 PCG76910.1 GAMC01015618 GAMC01015617 JAB90937.1 NEVH01002987 PNF41107.1 GAKP01002615 JAC56337.1 GDHF01014841 JAI37473.1 KQ971357 EFA09280.1 CCAG010005392 AJWK01016483 AJWK01016484 KK852699 KDR18213.1 CH916367 EDW00949.1 JXUM01081571 JXUM01081572 JXUM01081573 KQ563255 KXJ74195.1 CH933808 EDW09982.1 GFDF01002428 JAV11656.1 CH477254 EAT45917.1 EAT45918.1 JXUM01091697 JXUM01091698 JXUM01091699 KQ563935 KXJ73050.1 JRES01001254 KNC24206.1 CP012524 ALC40855.1 KA647170 KA649733 KA649753 AFP64362.1 CH963849 EDW74603.1 AJVK01009514 AJVK01009515 AJVK01009516 OUUW01000001 SPP75670.1 CM000071 EAL25351.1 CH479181 EDW32299.1 GALX01005629 JAB62837.1 GEZM01016729 JAV91162.1 AXCN02001095 CH902619 EDV36441.1 CM000157 EDW89182.1 CVRI01000040 CRK95020.1 CH954177 EDV59687.1 UFQT01000092 SSX19588.1 AXCM01000489 AMQN01008410 KB303059 ELU03519.1 AAAB01008986 EAA00329.3 CM002911 KMY91977.1 AF071103 AF071104 AE013599 AY122137 BT001560 BT001772 GFDL01005494 JAV29551.1 JR037601 AEY57868.1 KB200665 ESP00812.1 DS231941 EDS28093.1 GFDL01005495 JAV29550.1 ATLV01023946 KE525347 KFB50135.1 ADMH02000915 ETN64673.1 CH940648 EDW60990.1 GGLE01004683 MBY08809.1 KE163515 EPQ12449.1 GEBQ01000716 JAT39261.1 KK116827 KFM68884.1 JP011804 AES00402.1 KZ288478 PBC25266.1 KQ434870 KZC09470.1 GANO01002053 JAB57818.1 ADTU01003756 AEYP01051416 KQ414681 KOC63962.1 GL888331 EGI62697.1 GL436621 EFN71722.1 KQ981768 KYN36022.1 AAPE02040677 KQ976701 KYM77211.1 KQ435955 KOX68055.1 KB030270 ELK19191.1 RCHS01002306 RMX48175.1 GL451555 EFN79046.1 AANG04000016 GDIP01044160 JAM59555.1 GECU01011475 JAS96231.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000235965
UP000091820
+ More
UP000192223 UP000007266 UP000092443 UP000095300 UP000092444 UP000092461 UP000078200 UP000092445 UP000027135 UP000001070 UP000069940 UP000249989 UP000009192 UP000008820 UP000037069 UP000092553 UP000007798 UP000095301 UP000092462 UP000268350 UP000075881 UP000001819 UP000008744 UP000075884 UP000069272 UP000075886 UP000007801 UP000002282 UP000076408 UP000075903 UP000183832 UP000076407 UP000079169 UP000008711 UP000192221 UP000075883 UP000075900 UP000014760 UP000007062 UP000005203 UP000075882 UP000000803 UP000075902 UP000030746 UP000085678 UP000075885 UP000075920 UP000002320 UP000030765 UP000000673 UP000008792 UP000054359 UP000075880 UP000242457 UP000076502 UP000005205 UP000000715 UP000053825 UP000007755 UP000000311 UP000265180 UP000007879 UP000001038 UP000078541 UP000001074 UP000078540 UP000053105 UP000265200 UP000010552 UP000275408 UP000008237 UP000248482 UP000011712
UP000192223 UP000007266 UP000092443 UP000095300 UP000092444 UP000092461 UP000078200 UP000092445 UP000027135 UP000001070 UP000069940 UP000249989 UP000009192 UP000008820 UP000037069 UP000092553 UP000007798 UP000095301 UP000092462 UP000268350 UP000075881 UP000001819 UP000008744 UP000075884 UP000069272 UP000075886 UP000007801 UP000002282 UP000076408 UP000075903 UP000183832 UP000076407 UP000079169 UP000008711 UP000192221 UP000075883 UP000075900 UP000014760 UP000007062 UP000005203 UP000075882 UP000000803 UP000075902 UP000030746 UP000085678 UP000075885 UP000075920 UP000002320 UP000030765 UP000000673 UP000008792 UP000054359 UP000075880 UP000242457 UP000076502 UP000005205 UP000000715 UP000053825 UP000007755 UP000000311 UP000265180 UP000007879 UP000001038 UP000078541 UP000001074 UP000078540 UP000053105 UP000265200 UP000010552 UP000275408 UP000008237 UP000248482 UP000011712
Interpro
Gene 3D
ProteinModelPortal
H9JD44
A0A194QBG0
A0A194R539
A0A2A4JZS2
W8AXX6
A0A2J7RJU5
+ More
A0A1A9WR46 A0A1W4X2G4 A0A034WPS6 A0A0K8VF34 D6WV33 A0A1A9XFG4 A0A1I8PC50 A0A1B0G1H8 A0A1B0CKN3 A0A1A9V3A3 A0A1B0AGG2 A0A067R717 B4J6P6 A0A182GQX6 B4KNC9 A0A1L8DYZ6 Q17H06 A0A182GVS5 A0A0L0BW05 A0A0M4EIF0 T1PP12 B4MR64 A0A1I8NDV0 A0A1B0CZ62 A0A3B0JJW5 A0A182JXK2 Q290L0 B4GBP1 A0A182N3W5 A0A182FUZ8 V5GMK8 A0A1Y1MZR4 A0A182QAP8 B3MDQ6 A0A182HHE3 B4P1P6 A0A182YLA1 A0A182VF33 A0A1J1I609 A0A182XGS6 A0A1S4EBW8 B3N9Z1 A0A336M0U0 A0A1W4V0U6 A0A182MDN0 A0A182RYY1 R7UC15 Q7PZB9 A0A0J9R749 A0A088AHY9 A0A182L4Y4 O97477 A0A1Q3FPL3 V9I9S1 A0A182UEU2 V4B2T7 A0A1S3H4Z5 A0A182P7I1 A0A182W1X7 B0WI11 A0A1Q3FPM1 A0A084WIU1 W5JMX9 B4LMP9 A0A2R5LH56 S7N5Q4 A0A1B6MTK9 A0A087TUU5 A0A182J2U9 A0A1S3IQ99 G9K677 A0A2A3E1Y2 A0A154PE42 U5EV30 A0A158NYD0 M3YVS2 A0A0L7QZJ6 F4WSZ3 E2A4B2 A0A3P9KY71 A0A1X7V072 H2MGD4 A0A195F761 G1NX43 A0A195AYD6 A0A0M8ZNV0 A0A3P9HWT8 L5L7E3 A0A3M6U3B9 E2BZ44 A0A2Y9IYD6 M3WC06 A0A0P6ADI8 A0A1B6JAG6
A0A1A9WR46 A0A1W4X2G4 A0A034WPS6 A0A0K8VF34 D6WV33 A0A1A9XFG4 A0A1I8PC50 A0A1B0G1H8 A0A1B0CKN3 A0A1A9V3A3 A0A1B0AGG2 A0A067R717 B4J6P6 A0A182GQX6 B4KNC9 A0A1L8DYZ6 Q17H06 A0A182GVS5 A0A0L0BW05 A0A0M4EIF0 T1PP12 B4MR64 A0A1I8NDV0 A0A1B0CZ62 A0A3B0JJW5 A0A182JXK2 Q290L0 B4GBP1 A0A182N3W5 A0A182FUZ8 V5GMK8 A0A1Y1MZR4 A0A182QAP8 B3MDQ6 A0A182HHE3 B4P1P6 A0A182YLA1 A0A182VF33 A0A1J1I609 A0A182XGS6 A0A1S4EBW8 B3N9Z1 A0A336M0U0 A0A1W4V0U6 A0A182MDN0 A0A182RYY1 R7UC15 Q7PZB9 A0A0J9R749 A0A088AHY9 A0A182L4Y4 O97477 A0A1Q3FPL3 V9I9S1 A0A182UEU2 V4B2T7 A0A1S3H4Z5 A0A182P7I1 A0A182W1X7 B0WI11 A0A1Q3FPM1 A0A084WIU1 W5JMX9 B4LMP9 A0A2R5LH56 S7N5Q4 A0A1B6MTK9 A0A087TUU5 A0A182J2U9 A0A1S3IQ99 G9K677 A0A2A3E1Y2 A0A154PE42 U5EV30 A0A158NYD0 M3YVS2 A0A0L7QZJ6 F4WSZ3 E2A4B2 A0A3P9KY71 A0A1X7V072 H2MGD4 A0A195F761 G1NX43 A0A195AYD6 A0A0M8ZNV0 A0A3P9HWT8 L5L7E3 A0A3M6U3B9 E2BZ44 A0A2Y9IYD6 M3WC06 A0A0P6ADI8 A0A1B6JAG6
PDB
1RM0
E-value=3.82012e-141,
Score=1287
Ontologies
PATHWAY
GO
PANTHER
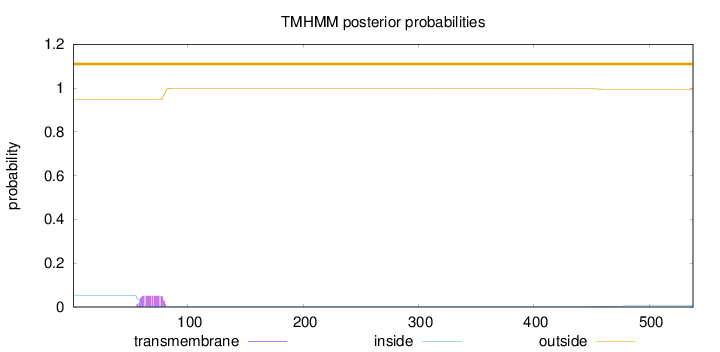
Topology
Subcellular location
Cytoplasm
Length:
538
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.22721
Exp number, first 60 AAs:
0.13722
Total prob of N-in:
0.05201
outside
1 - 538
Population Genetic Test Statistics
Pi
181.952662
Theta
163.23499
Tajima's D
1.196152
CLR
0.179569
CSRT
0.710964451777411
Interpretation
Uncertain
