Gene
KWMTBOMO01122
Pre Gene Modal
BGIBMGA007330
Annotation
PREDICTED:_sex_determination_protein_fruitless-like_isoform_X3_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.723
Sequence
CDS
ATGCAGGGGCTGGTGAGCGACAAGACGTTCCACTTGAAGTGGAACAACCACCTGCAGAACCTGAGCCAGCTGTTTACGACCATCTACTCGTCGTCAGCGCTGGCGGACGTCACGCTCTCCTGCCGCGATGGGACCCTCAAGGCGCACAAACTTGTGCTCTCAGCTTGTAGTCCGTATTTCGAACAAATATTCAAGGACAATCCATGTCAGCACCCCATCGTGATTTTAAAAGGGATCCCTTTTTCTGAGATCAACCTCCTTGTGGAATTCATGTACAAAGGGTCAGTGGATGTTCAAGAATTGGACCTGCAGTCATTAATGCATACAGCTTCCGAGTTAGAGATCCGAGGACTAGCTTATGAAGCTCGTGACAATGCTGCCCAGCTTTTAAATGTGAATTTAGAGTACCCCACAACATATCCACAGACAGCAACTTCCACTCAATCCTCGTTTCAACAAGCTCGTGCAACAGACGTTGAAAGACTAAAACAGATACACATGTACCAGCAATCAATGATGCGTGCCGAAGAGATTAGGCGGAGGCGTCGCGACGACAACCCTGGAACGCACACGGCGGTCAATAATATTTTAGCTGCTGCAGCCAGGGAAATGGAAGAGGCTAGGAATGCTGCAAGAGTCAGCGAACGCGTCGCTGCCACTATACAGACGGAGCACGAGCAGGCTCTCGCAGCGGCCACGTCAACGACACCAGAACTAAAGCGTGAATCTGAAGACGATCGCCCCAAAAAACGGGTCTCGATTTTGTCACCGGAAGACGACAAAATGAGGGCCAAGAAGAAACCTATGACAGTACGTTTCCAAGACGAGAAAGCTGACGAAAGATCCAGCAAAGTGACTTCACCCGGGGCGGCCAGTAATGATTCGTTGAACAAGAACGACACTACGAGCGGTATTAAGGTGGTGCAACTCTCGATGCTACAGAAGCCCCCGAACAACGAACACGACGACTCCGACGAGAAGTCTGGTCACAAAGGCGTGATGGACGACGACGAGGAAACAGATTCGAGCGGTTCGATGTCGGATTCGGCCGCGAACGACGCGCAGGACGGGCGCCCGCACGTCTGCGACGTGTGCGATCTGAGATTCCAAAGGTCATCGCATCTGAGCCGCCACAAGCTGACCCACACGGGCGAGCGACCCTACACGTGCGGCGGCTGCGGGAGGTCCTTCGCTAGATCTGACAAGCTACGAATGCACACCAAGATCTGTGAGAGGGAGGACCCAACGGTGGACATGACAAACGACGCGCCGTCGAATAAACGCGAGAGCAACTCTGAAGTGATGTCCGGCATGGTGCGGAGCACGCACCGCGAGTCCGGCCACCTGGTGTTCACGCCGGGCGGTGACGTCATGCTGCCCCCCCGCCCCGCGCCCGCCGTGCTGGAGCGCGCCGACCCCCCGCCCGACGGCATGCTCGACCCCCCGCGGAGGGGACGCGGCCGGCCCAGGAAGACTCCTCTGCCTCTAACGCCTAAAATCAAGAAGAAGAGAGGTCGTCCCCCTAAGACATCGCTCGACCTGGAAGGTTGCGTACTAACGAGCGGCTCAGTGATGGGTCCGAGCCCTCAGCAGATCTCCCACATCACGACTCCCTCCGGACACCAACTGTTACTCAAGAGGAAACGTGGACGACCTCCGAAGAATTATTTCATGCCCAAACAAGATGGACGTCCGAACGAAAACTACAATGCGACTAATCTACCCTTCGGCGACTTCTCGTACCTGACGGAAATGATCTACAACCCCATGACGTACAGCTACGGCGTGACCAGCGTCGACCCGGACCGGAACGTCGACACCGACCAGAGCATGTCCACCGAGCCGCACGACCGGACCATCGACGTGTCCGACACGTCATCGGACGACGACGACTCGCGAGTCGAGCCGGAAGAGAACATCCCTGCCGTTGCCATGGAAACGCAGATCATGACCGTCGGCGACTGTCAGATCGTCAAACTTCCCGCCAACAACGATGATAAGGACGACAAAATTGATCAATCGGCTGAGGGTAGTGTAGTGACGTCATCGACTCCTAGCTCGGTCACCATCACCCCTATAACCACAGTGGGCGACTGCACCATACGGCCCGTCGAGAACACATAA
Protein
MQGLVSDKTFHLKWNNHLQNLSQLFTTIYSSSALADVTLSCRDGTLKAHKLVLSACSPYFEQIFKDNPCQHPIVILKGIPFSEINLLVEFMYKGSVDVQELDLQSLMHTASELEIRGLAYEARDNAAQLLNVNLEYPTTYPQTATSTQSSFQQARATDVERLKQIHMYQQSMMRAEEIRRRRRDDNPGTHTAVNNILAAAAREMEEARNAARVSERVAATIQTEHEQALAAATSTTPELKRESEDDRPKKRVSILSPEDDKMRAKKKPMTVRFQDEKADERSSKVTSPGAASNDSLNKNDTTSGIKVVQLSMLQKPPNNEHDDSDEKSGHKGVMDDDEETDSSGSMSDSAANDAQDGRPHVCDVCDLRFQRSSHLSRHKLTHTGERPYTCGGCGRSFARSDKLRMHTKICEREDPTVDMTNDAPSNKRESNSEVMSGMVRSTHRESGHLVFTPGGDVMLPPRPAPAVLERADPPPDGMLDPPRRGRGRPRKTPLPLTPKIKKKRGRPPKTSLDLEGCVLTSGSVMGPSPQQISHITTPSGHQLLLKRKRGRPPKNYFMPKQDGRPNENYNATNLPFGDFSYLTEMIYNPMTYSYGVTSVDPDRNVDTDQSMSTEPHDRTIDVSDTSSDDDDSRVEPEENIPAVAMETQIMTVGDCQIVKLPANNDDKDDKIDQSAEGSVVTSSTPSSVTITPITTVGDCTIRPVENT
Summary
Pubmed
EMBL
Proteomes
Interpro
ProteinModelPortal
PDB
1A1H
E-value=4.06766e-10,
Score=158
Ontologies
GO
Topology
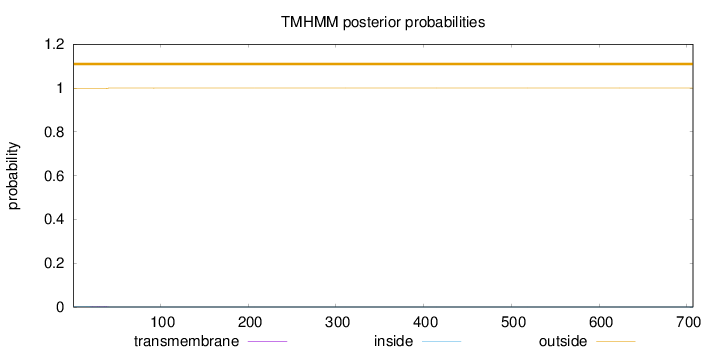
Length:
707
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0373900000000001
Exp number, first 60 AAs:
0.03497
Total prob of N-in:
0.00197
outside
1 - 707
Population Genetic Test Statistics
Pi
206.348135
Theta
160.83584
Tajima's D
0.591874
CLR
1.21479
CSRT
0.540422978851058
Interpretation
Uncertain
