Gene
KWMTBOMO01116
Pre Gene Modal
BGIBMGA007331
Annotation
PREDICTED:_protein_MCM10_homolog_isoform_X1_[Bombyx_mori]
Full name
Protein MCM10 homolog
Alternative Name
Sensitized chromosome inheritance modifier 19
Location in the cell
Nuclear Reliability : 4.496
Sequence
CDS
ATGGACGAGAAAGATGAATTATCACAGCTTGAAGAACTGCTCAGTGCTGACTTCGGTGAAAATACTGAAATGAATGCACAAAATAACACCAAAGAAGTAGACATATTTCAGCAAACTACAAACTTAAGAAGTGAAATGCCATCATCAAGTACAGCTGAAGTTCCTTCTATTGTACACAATGGAGATACTGATTCTTCAGACGACGAGGAAAAGCGGGACTATGCAGAAAGGAAGTACAATGATTACGGTTCACAAGTAAAAAAATCATTAGACAAAATTGAACAAGATCAAATTAATCAAAGACTGGATTTTGAGAGTAGATTACGACAAACAAACATGGAAACTAAAGTTTCACGTTATTCCAAATTCACAAAAAAACCTAACATTGAACCTAGAAATGTGTTCAGTCCTCCTAAAGTGAAAAGTAATAAAAATGCAGATGTTTACACAGACCCTATATTCAATATTAGGATTACAAAGCCATTGCTATCTAGTTCAGTTTTGCTAGAGAGAATGCAAGGTAGAGAAGCTATAAGCATGCTTAGAGTAAAAAGGCATGTTGAGAATGAAGACTTGTCAAAAGATTGGGTGATTGCAGGTGTTATTGTTAGGAAAAGTTCAACTAAACAATCTCAGAAAGGAAGTCAGTTTATTATATGGACTCTCAGTGATTTAAAAGATGATATGAAAACGGTTTCTATATTTTTATTTAGGAAAGCCTATAATGAACTATGGAAAACTGCTGAAGGAACAGTTGTTGCAGTTCTTAACCCTAATGTATTAGATAGATCACAGAATAGCGCAGATCAGGCTTGCCTCAGTGTTGATAATCCTGACAGAGTTATGATCATAGGACTCTCCAAAGATTTTGGTATTTGTAAAAGTAAGAAGAAAAATGGGGAACCATGCACAGCCTTTGTCAATATGAATCAATGTCAACACTGCATTTATCATGTAAAACAAGAATACCAAAAGTTTTCACAGAGACAAGAACTACAATCTTCAACCATGGGCAAGGGTTTAGTAAGTTTACAAAATAAAGTTTTAGGAAAAAACACTGTGATCTATGGAGGCAAAGCATATTCAGCCTTACAAAGCGGAAATAACAAAAAGATAAAAGAAAAAGATCAAAACAGACTGATGTCACTCAGTGACTATTACAAAACTGAGGGTGACAAGTTAATCACTAACAAAATGCCTTATGGGGCAGGATTTGTGAAGAACAATAGTGCATTACACAGTCCGACTACACAAAAAACTAGTGATACTGAAAGACTCAGCAAATTGTCCAACTTTAATACACCTACAAACTCAAAACAACTTAGCCTTGCAGAAAAACTGACTAATTCTCCCACATTAGGCAAAGGATTTAGTATAAAAGGCAGTGGAACAATAGACCTAAATATATCATACAAACAAAAGGTAGCTGAAAAAGCAAAATCCAATGCAATCAGAATAATTCAACAGAATGGTGGATTACCAAAATTAGATCCTAATAATTTAAATGGAACTGCTGCTGGGAAAAAAAGAGCTTTAGAAAAATTGAATGGAATGAATGATGAGAATAGCAGTAAAAAAGCAAAATCCAATCATGAACAGAGCAACACTGGGAAAATACTCTCTGAAAGGTTCCGTAAAATATTAGAAGCCACATCAGTACATCAAGACTTAGTAAAACAACATGAAGATGATGAACAAGAGAAGTATTTTAACAAATTAGAGAAAAAGGAAGCAATGGAAGAAAAAATGTTGAGCACATATAAACTTCCATGCAAAGCAGTAAGATGTGCTAAATGTAAATACACTGCATTTTCTGCTGCTCAACGGTGTAAAGATGAAAGACATCCTCTAAAAGTATTAGATACATTTAAGAGGTTCTTTAAATGTTTGGATTGTAAAAATAGAACTGTTTCTTTAGAATTGCTGCCTCTGCATTCTTGTAATAATTGTGGAAGTTCACGGTGGGAGAAAGCTCCAATGTTAAGAGAAAAGAAAATAACGACACTTTCTGATGGTTTATCCATCAGGGGTGAAGAAGAAACATTTTTGGGTGGACAAGTTGCTACTGGCAAAAATATAAATCTATTGGTACCAGACAGCTAA
Protein
MDEKDELSQLEELLSADFGENTEMNAQNNTKEVDIFQQTTNLRSEMPSSSTAEVPSIVHNGDTDSSDDEEKRDYAERKYNDYGSQVKKSLDKIEQDQINQRLDFESRLRQTNMETKVSRYSKFTKKPNIEPRNVFSPPKVKSNKNADVYTDPIFNIRITKPLLSSSVLLERMQGREAISMLRVKRHVENEDLSKDWVIAGVIVRKSSTKQSQKGSQFIIWTLSDLKDDMKTVSIFLFRKAYNELWKTAEGTVVAVLNPNVLDRSQNSADQACLSVDNPDRVMIIGLSKDFGICKSKKKNGEPCTAFVNMNQCQHCIYHVKQEYQKFSQRQELQSSTMGKGLVSLQNKVLGKNTVIYGGKAYSALQSGNNKKIKEKDQNRLMSLSDYYKTEGDKLITNKMPYGAGFVKNNSALHSPTTQKTSDTERLSKLSNFNTPTNSKQLSLAEKLTNSPTLGKGFSIKGSGTIDLNISYKQKVAEKAKSNAIRIIQQNGGLPKLDPNNLNGTAAGKKRALEKLNGMNDENSSKKAKSNHEQSNTGKILSERFRKILEATSVHQDLVKQHEDDEQEKYFNKLEKKEAMEEKMLSTYKLPCKAVRCAKCKYTAFSAAQRCKDERHPLKVLDTFKRFFKCLDCKNRTVSLELLPLHSCNNCGSSRWEKAPMLREKKITTLSDGLSIRGEEETFLGGQVATGKNINLLVPDS
Summary
Description
Proposed to be involved in DNA replication and to participate in the activation of the pre-replication complex (pre-RC). May be involved in chromosome condensation.
Subunit
Self-associates. Interacts with Mcm2, dup, Orc2, CDC45L and Su(var)205.
Similarity
Belongs to the MCM10 family.
Keywords
Complete proteome
DNA replication
Metal-binding
Nucleus
Phosphoprotein
Reference proteome
Zinc
Zinc-finger
Feature
chain Protein MCM10 homolog
Uniprot
A0A0L7KYK6
A0A2H1W6P2
A0A2W1B2R1
A0A0N1PGC4
A0A194QBH0
A0A2A4JZU4
+ More
A0A212F5Y5 A0A336M324 A0A1Q3EWW6 B0W0E0 A0A182GCN5 B0XLH7 Q17D82 A0A1S4F772 A0A336MX92 B4N7B8 A0A1W4UXI8 B3MN70 A0A1I8PQP2 A0A182JWH1 B4GJJ9 Q29NT4 B4IFM1 A0A3B0JMS4 A9UNF9 Q9VIE6 B4JAE6 A0A1I8M363 A0A182NAN1 A0A1Y1K9Q4 B4Q403 B4LU06 B4KF43 A0A1A9UPS9 A0A067RAI8 A0A182R410 A0A084VMI5 A0A0L0CB14 A0A1B0A5T7 A0A1A9Y0C4 B4P6Y2 A0A182JEC0 B3NKT1 A0A182P6F7 A0A1B0FPB2 A0A1B0CH79 A0A3L8DIM1 A0A1B0AS88 A0A0M3QU63 A0A182X691 A0A182IBB1 A0A195FTB8 A0A182KWG2 A0A182VFJ8 A0A182U1B4 A0A151WX28 A0A195DKJ0 F4WBG6 A0A1W4XAW2 A0A158P125 A0A195B3F0 A0A182F3R8 E2BQS1 E2A5U9 A0A0J7KJK8 A0A2R7VPG1 A0A182Q2M5 A0A1J1ING7 A0A026X0J5 A0A0L7QNG6 A0A1W4XAM4 A0A154PQ73 A0A087ZZG6 A0A232EU18 A0A0M9A4X7 A0A2A3EMU5 K7IXJ7 A0A0C9RRY7 A0A336N3B0 A0A310SJL9 T1I9G7 W8BT57 A0A0A1XHM1 A0A0C9RUP4 A0A0K8U4Y0 A0A034VS51 A0A2S2QIN5 E0VMQ2 J9K4B0 E9G6I1 A0A0P4YY76 A0A0P5VC16 A0A0P5ZUC8
A0A212F5Y5 A0A336M324 A0A1Q3EWW6 B0W0E0 A0A182GCN5 B0XLH7 Q17D82 A0A1S4F772 A0A336MX92 B4N7B8 A0A1W4UXI8 B3MN70 A0A1I8PQP2 A0A182JWH1 B4GJJ9 Q29NT4 B4IFM1 A0A3B0JMS4 A9UNF9 Q9VIE6 B4JAE6 A0A1I8M363 A0A182NAN1 A0A1Y1K9Q4 B4Q403 B4LU06 B4KF43 A0A1A9UPS9 A0A067RAI8 A0A182R410 A0A084VMI5 A0A0L0CB14 A0A1B0A5T7 A0A1A9Y0C4 B4P6Y2 A0A182JEC0 B3NKT1 A0A182P6F7 A0A1B0FPB2 A0A1B0CH79 A0A3L8DIM1 A0A1B0AS88 A0A0M3QU63 A0A182X691 A0A182IBB1 A0A195FTB8 A0A182KWG2 A0A182VFJ8 A0A182U1B4 A0A151WX28 A0A195DKJ0 F4WBG6 A0A1W4XAW2 A0A158P125 A0A195B3F0 A0A182F3R8 E2BQS1 E2A5U9 A0A0J7KJK8 A0A2R7VPG1 A0A182Q2M5 A0A1J1ING7 A0A026X0J5 A0A0L7QNG6 A0A1W4XAM4 A0A154PQ73 A0A087ZZG6 A0A232EU18 A0A0M9A4X7 A0A2A3EMU5 K7IXJ7 A0A0C9RRY7 A0A336N3B0 A0A310SJL9 T1I9G7 W8BT57 A0A0A1XHM1 A0A0C9RUP4 A0A0K8U4Y0 A0A034VS51 A0A2S2QIN5 E0VMQ2 J9K4B0 E9G6I1 A0A0P4YY76 A0A0P5VC16 A0A0P5ZUC8
Pubmed
26227816
28756777
26354079
22118469
26483478
17510324
+ More
17994087 15632085 23185243 10731132 12537572 12537569 12808023 18327897 25315136 28004739 22936249 24845553 24438588 26108605 17550304 30249741 20966253 21719571 21347285 20798317 24508170 28648823 20075255 24495485 25830018 25348373 20566863 21292972
17994087 15632085 23185243 10731132 12537572 12537569 12808023 18327897 25315136 28004739 22936249 24845553 24438588 26108605 17550304 30249741 20966253 21719571 21347285 20798317 24508170 28648823 20075255 24495485 25830018 25348373 20566863 21292972
EMBL
JTDY01004465
KOB68124.1
ODYU01006615
SOQ48616.1
KZ150559
PZC70538.1
+ More
KQ460468 KPJ14589.1 KQ459249 KPJ02335.1 NWSH01000370 PCG76922.1 AGBW02010127 OWR49119.1 UFQT01000336 SSX23329.1 GFDL01015236 JAV19809.1 DS231817 EDS39679.1 JXUM01160059 KQ573563 KXJ67921.1 DS234473 EDS34138.1 CH477298 EAT44347.1 UFQS01003243 UFQT01003243 SSX15421.1 SSX34790.1 CH964182 EDW80259.1 CH902620 EDV31027.1 CH479184 EDW36815.1 CH379058 EAL34560.1 KRT03652.1 CH480833 EDW46443.1 OUUW01000006 SPP82163.1 BT031323 ABY21736.1 AE014134 AY061477 CH916368 EDW03817.1 GEZM01095141 JAV55487.1 CM000361 CM002910 EDX05719.1 KMY91300.1 CH940649 EDW65059.1 CH933807 EDW13026.1 KK852819 KDR15628.1 ATLV01014596 KE524975 KFB39179.1 JRES01000668 KNC29410.1 CM000158 EDW89951.1 CH954179 EDV54454.1 CCAG010016105 AJWK01012087 QOIP01000008 RLU19688.1 JXJN01002691 CP012523 ALC40113.1 APCN01000664 KQ981272 KYN43855.1 KQ982691 KYQ52215.1 KQ980765 KYN13366.1 GL888063 EGI68445.1 ADTU01006100 KQ976641 KYM78807.1 GL449799 EFN81946.1 GL437061 EFN71191.1 LBMM01006623 KMQ90452.1 KK854016 PTY09412.1 AXCN02000614 CVRI01000054 CRL00638.1 KK107064 EZA60919.1 KQ414855 KOC60109.1 KQ435034 KZC14053.1 NNAY01002185 OXU21855.1 KQ435732 KOX77320.1 KZ288212 PBC32814.1 AAZX01004642 GBYB01011340 JAG81107.1 UFQS01003905 UFQT01003905 SSX16024.1 SSX35357.1 KQ761906 OAD56463.1 ACPB03017551 GAMC01014173 GAMC01014171 JAB92384.1 GBXI01003835 JAD10457.1 GBYB01011341 JAG81108.1 GDHF01030914 JAI21400.1 GAKP01014564 JAC44388.1 GGMS01008385 MBY77588.1 AAZO01003684 DS235321 EEB14658.1 ABLF02025674 GL732533 EFX84989.1 GDIP01221876 JAJ01526.1 GDIP01102388 JAM01327.1 GDIP01038996 LRGB01002384 JAM64719.1 KZS07922.1
KQ460468 KPJ14589.1 KQ459249 KPJ02335.1 NWSH01000370 PCG76922.1 AGBW02010127 OWR49119.1 UFQT01000336 SSX23329.1 GFDL01015236 JAV19809.1 DS231817 EDS39679.1 JXUM01160059 KQ573563 KXJ67921.1 DS234473 EDS34138.1 CH477298 EAT44347.1 UFQS01003243 UFQT01003243 SSX15421.1 SSX34790.1 CH964182 EDW80259.1 CH902620 EDV31027.1 CH479184 EDW36815.1 CH379058 EAL34560.1 KRT03652.1 CH480833 EDW46443.1 OUUW01000006 SPP82163.1 BT031323 ABY21736.1 AE014134 AY061477 CH916368 EDW03817.1 GEZM01095141 JAV55487.1 CM000361 CM002910 EDX05719.1 KMY91300.1 CH940649 EDW65059.1 CH933807 EDW13026.1 KK852819 KDR15628.1 ATLV01014596 KE524975 KFB39179.1 JRES01000668 KNC29410.1 CM000158 EDW89951.1 CH954179 EDV54454.1 CCAG010016105 AJWK01012087 QOIP01000008 RLU19688.1 JXJN01002691 CP012523 ALC40113.1 APCN01000664 KQ981272 KYN43855.1 KQ982691 KYQ52215.1 KQ980765 KYN13366.1 GL888063 EGI68445.1 ADTU01006100 KQ976641 KYM78807.1 GL449799 EFN81946.1 GL437061 EFN71191.1 LBMM01006623 KMQ90452.1 KK854016 PTY09412.1 AXCN02000614 CVRI01000054 CRL00638.1 KK107064 EZA60919.1 KQ414855 KOC60109.1 KQ435034 KZC14053.1 NNAY01002185 OXU21855.1 KQ435732 KOX77320.1 KZ288212 PBC32814.1 AAZX01004642 GBYB01011340 JAG81107.1 UFQS01003905 UFQT01003905 SSX16024.1 SSX35357.1 KQ761906 OAD56463.1 ACPB03017551 GAMC01014173 GAMC01014171 JAB92384.1 GBXI01003835 JAD10457.1 GBYB01011341 JAG81108.1 GDHF01030914 JAI21400.1 GAKP01014564 JAC44388.1 GGMS01008385 MBY77588.1 AAZO01003684 DS235321 EEB14658.1 ABLF02025674 GL732533 EFX84989.1 GDIP01221876 JAJ01526.1 GDIP01102388 JAM01327.1 GDIP01038996 LRGB01002384 JAM64719.1 KZS07922.1
Proteomes
UP000037510
UP000053240
UP000053268
UP000218220
UP000007151
UP000002320
+ More
UP000069940 UP000249989 UP000008820 UP000007798 UP000192221 UP000007801 UP000095300 UP000075881 UP000008744 UP000001819 UP000001292 UP000268350 UP000000803 UP000001070 UP000095301 UP000075884 UP000000304 UP000008792 UP000009192 UP000078200 UP000027135 UP000075900 UP000030765 UP000037069 UP000092445 UP000092443 UP000002282 UP000075880 UP000008711 UP000075885 UP000092444 UP000092461 UP000279307 UP000092460 UP000092553 UP000076407 UP000075840 UP000078541 UP000075882 UP000075903 UP000075902 UP000075809 UP000078492 UP000007755 UP000192223 UP000005205 UP000078540 UP000069272 UP000008237 UP000000311 UP000036403 UP000075886 UP000183832 UP000053097 UP000053825 UP000076502 UP000005203 UP000215335 UP000053105 UP000242457 UP000002358 UP000015103 UP000009046 UP000007819 UP000000305 UP000076858
UP000069940 UP000249989 UP000008820 UP000007798 UP000192221 UP000007801 UP000095300 UP000075881 UP000008744 UP000001819 UP000001292 UP000268350 UP000000803 UP000001070 UP000095301 UP000075884 UP000000304 UP000008792 UP000009192 UP000078200 UP000027135 UP000075900 UP000030765 UP000037069 UP000092445 UP000092443 UP000002282 UP000075880 UP000008711 UP000075885 UP000092444 UP000092461 UP000279307 UP000092460 UP000092553 UP000076407 UP000075840 UP000078541 UP000075882 UP000075903 UP000075902 UP000075809 UP000078492 UP000007755 UP000192223 UP000005205 UP000078540 UP000069272 UP000008237 UP000000311 UP000036403 UP000075886 UP000183832 UP000053097 UP000053825 UP000076502 UP000005203 UP000215335 UP000053105 UP000242457 UP000002358 UP000015103 UP000009046 UP000007819 UP000000305 UP000076858
ProteinModelPortal
A0A0L7KYK6
A0A2H1W6P2
A0A2W1B2R1
A0A0N1PGC4
A0A194QBH0
A0A2A4JZU4
+ More
A0A212F5Y5 A0A336M324 A0A1Q3EWW6 B0W0E0 A0A182GCN5 B0XLH7 Q17D82 A0A1S4F772 A0A336MX92 B4N7B8 A0A1W4UXI8 B3MN70 A0A1I8PQP2 A0A182JWH1 B4GJJ9 Q29NT4 B4IFM1 A0A3B0JMS4 A9UNF9 Q9VIE6 B4JAE6 A0A1I8M363 A0A182NAN1 A0A1Y1K9Q4 B4Q403 B4LU06 B4KF43 A0A1A9UPS9 A0A067RAI8 A0A182R410 A0A084VMI5 A0A0L0CB14 A0A1B0A5T7 A0A1A9Y0C4 B4P6Y2 A0A182JEC0 B3NKT1 A0A182P6F7 A0A1B0FPB2 A0A1B0CH79 A0A3L8DIM1 A0A1B0AS88 A0A0M3QU63 A0A182X691 A0A182IBB1 A0A195FTB8 A0A182KWG2 A0A182VFJ8 A0A182U1B4 A0A151WX28 A0A195DKJ0 F4WBG6 A0A1W4XAW2 A0A158P125 A0A195B3F0 A0A182F3R8 E2BQS1 E2A5U9 A0A0J7KJK8 A0A2R7VPG1 A0A182Q2M5 A0A1J1ING7 A0A026X0J5 A0A0L7QNG6 A0A1W4XAM4 A0A154PQ73 A0A087ZZG6 A0A232EU18 A0A0M9A4X7 A0A2A3EMU5 K7IXJ7 A0A0C9RRY7 A0A336N3B0 A0A310SJL9 T1I9G7 W8BT57 A0A0A1XHM1 A0A0C9RUP4 A0A0K8U4Y0 A0A034VS51 A0A2S2QIN5 E0VMQ2 J9K4B0 E9G6I1 A0A0P4YY76 A0A0P5VC16 A0A0P5ZUC8
A0A212F5Y5 A0A336M324 A0A1Q3EWW6 B0W0E0 A0A182GCN5 B0XLH7 Q17D82 A0A1S4F772 A0A336MX92 B4N7B8 A0A1W4UXI8 B3MN70 A0A1I8PQP2 A0A182JWH1 B4GJJ9 Q29NT4 B4IFM1 A0A3B0JMS4 A9UNF9 Q9VIE6 B4JAE6 A0A1I8M363 A0A182NAN1 A0A1Y1K9Q4 B4Q403 B4LU06 B4KF43 A0A1A9UPS9 A0A067RAI8 A0A182R410 A0A084VMI5 A0A0L0CB14 A0A1B0A5T7 A0A1A9Y0C4 B4P6Y2 A0A182JEC0 B3NKT1 A0A182P6F7 A0A1B0FPB2 A0A1B0CH79 A0A3L8DIM1 A0A1B0AS88 A0A0M3QU63 A0A182X691 A0A182IBB1 A0A195FTB8 A0A182KWG2 A0A182VFJ8 A0A182U1B4 A0A151WX28 A0A195DKJ0 F4WBG6 A0A1W4XAW2 A0A158P125 A0A195B3F0 A0A182F3R8 E2BQS1 E2A5U9 A0A0J7KJK8 A0A2R7VPG1 A0A182Q2M5 A0A1J1ING7 A0A026X0J5 A0A0L7QNG6 A0A1W4XAM4 A0A154PQ73 A0A087ZZG6 A0A232EU18 A0A0M9A4X7 A0A2A3EMU5 K7IXJ7 A0A0C9RRY7 A0A336N3B0 A0A310SJL9 T1I9G7 W8BT57 A0A0A1XHM1 A0A0C9RUP4 A0A0K8U4Y0 A0A034VS51 A0A2S2QIN5 E0VMQ2 J9K4B0 E9G6I1 A0A0P4YY76 A0A0P5VC16 A0A0P5ZUC8
PDB
3EBE
E-value=9.49538e-38,
Score=396
Ontologies
GO
PANTHER
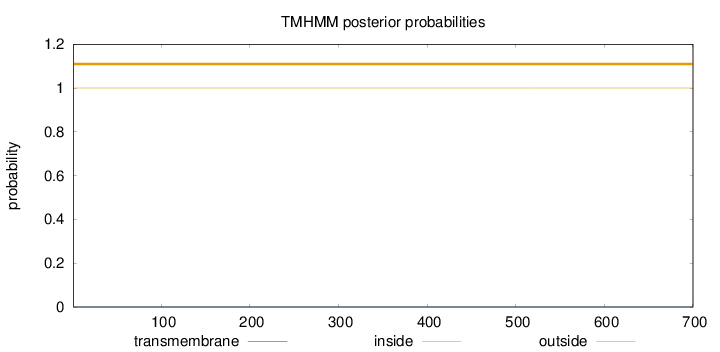
Topology
Subcellular location
Nucleus
Length:
700
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00011
outside
1 - 700
Population Genetic Test Statistics
Pi
167.90158
Theta
163.329778
Tajima's D
-0.154978
CLR
51.038418
CSRT
0.330633468326584
Interpretation
Uncertain
