Gene
KWMTBOMO01115 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007436
Annotation
PREDICTED:_GMP_synthase_[glutamine-hydrolyzing]_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 2.197
Sequence
CDS
ATGCACACAGCGAGTGCGGACGGACAGTTCCGCGGCACCAATGGCTCCGGCCAGGGACGTGACAAAGTTGCCATACTGGACGCCGGCTCACAATATGGGAAGGTGATAGACCGCCGCGTGCGCGAGCTGTGCGTCGAATCGGAGATCCTACCGTTGGAGACCTCGGCCTACCAGCTGAAGGAGGCCGGATACCGCGCCATCATCATTTCGGGCGGACCCAACTCTGTGTACGCCGAGAATGCGCCGAAATACGACGCGGACGTCTTCAAAATCGGCCTTCCGGTTTTAGGCATATGTTATGGGATGCAAATGTTGAACAAGGAGTTTGGTGGCTCCGTGCTGCGTAAGGACGCGCGCGAGGACGGGCAGTACGAGATCGAAGTGGAAACCAGCTGCCCTTTGTTCAATCGTCTGGAGAAGCAGCAGCCGGTGCTCCTGACCCACGGAGACAGCGTCCACAAGGTGGGAGACAGGTTCCGTGTGGCCGCGCAGTCTTCGAACCACCTTATCGCGGCCATATACAACGAGCAGATGAAGCTGTACGGGGTGCAGTTCCATCCCGAGGTGGATCTAACCCCTAAAGGCAAGCAGATGTTGTCCAACTTCCTGTTCGACATCGCCGGCCTCTCGCGCTCCTTCACGCTGCGTTGTCGGCGCGAGGCCTGCGTGCAGTACATCAAGGAGACCGTCGGCGAAAACAATAAAGTTCTGGTGTTGGTGAGTGGCGGTGTGGATTCAACGGTATGTGCAGCCTTGCTGAGGACAGTGCTTCGTGAGGATCAGGTCATAGCACTACACATTGATAATGGTTTTATGCGTAAGAACGAAAGTGCCAAAGTTGAGAGGTGCCTCCGTGAGTTGGGAGTCCGCCTGCACGTCATTAACGCCGCTCATCAGTTCTTTCACGGCAGCACAGTGGTCCGCGAGTCAGGAGGCTGGTCCCGCCACGCCGGGCCGCTGTGTCACACCGCCTCGCCGGAGGACAAGCGGAAGATCATAGGAGATGTATTCATACGGATCGCGGAACAGGCCGTCCACGAACTTAATCTGCAAGAGGACGAAGTACTTCTAGGGCAGGGCACGCTGCGCCCCGACCTCATCGAGTCCGCGTCGGCGCTGTGCTCGGGCCAGGCCGACACCATCAAGACGCACCACAACGACACCGAGCTGGTGCGGGCGCTGCGGCAGAGGGGACGGGTCGTGGAGCCTCTGAAGGACTTCCATAAAGACGAGGTCCGAGCGCTGGGGATGGAGCTGGGGCTGCCCGAGGCGCTGGTGGAGCGGCACCCGTTCCCGGGCCCCGGCCTCGCCGTCAGGGTCATATGTCAAGACGAACCGTTCATGGAGAGGGACTTCGCCGAAACGCAGGTGATAGTAAAAATAATAGTGGAGTTCGCATCGATGTACCTCAAGTCCCACGCGCTGCTGGGCCGCGTCGTGAACGCGAGCACGCCCGCCGAGCACGCCAAGCTGCGCCGCATCTCGTCCGCATACCAGGTCGCCGCCACGCTGCTGCCCATCAGAACGGTCGGAGTGCAAGGTGATTGCAGAACATACAGCTATGCCGTGGCATTGTCAACGGAGAAGTATCCTCCGGATTGGAAAGACATGCAGTATCTCGCTCAAATAATACCGAGGATCTGTCATAATGTTAACAGGGTATGTTACGCTTTTGGAGGTCTGATCAAGGAGCAAGTAACAGATATCACACCAACTTTCCTTACGCAACAAGTTGTTTCAACTTTACGACAGGCCGACGACCTGGCAACACAGGTGCTGACCCAGAGCGGATGCGCCGGCCGCATCGCACAGATGCCGGTGGTGCTGATCCCGATCCAGTTCGACCGCGACGCCGCGGTGCGCGCCGCCTCGTGCCAGCGGTCTATCGTACTGCGTCCTTTCGTCACGTCAGACTTCATGACCGGTCTCGCCGCACTCCCGGGATCCGACCACATGCCTCAAGACGTTGTGGACAGAATGTGTAAGAAGCTCTTGACCGTGCCTGGAATATCTCGCGTATTATACGACCTAACACCGAAACCGCCCGCCACAACAGAATGGGAATGA
Protein
MHTASADGQFRGTNGSGQGRDKVAILDAGSQYGKVIDRRVRELCVESEILPLETSAYQLKEAGYRAIIISGGPNSVYAENAPKYDADVFKIGLPVLGICYGMQMLNKEFGGSVLRKDAREDGQYEIEVETSCPLFNRLEKQQPVLLTHGDSVHKVGDRFRVAAQSSNHLIAAIYNEQMKLYGVQFHPEVDLTPKGKQMLSNFLFDIAGLSRSFTLRCRREACVQYIKETVGENNKVLVLVSGGVDSTVCAALLRTVLREDQVIALHIDNGFMRKNESAKVERCLRELGVRLHVINAAHQFFHGSTVVRESGGWSRHAGPLCHTASPEDKRKIIGDVFIRIAEQAVHELNLQEDEVLLGQGTLRPDLIESASALCSGQADTIKTHHNDTELVRALRQRGRVVEPLKDFHKDEVRALGMELGLPEALVERHPFPGPGLAVRVICQDEPFMERDFAETQVIVKIIVEFASMYLKSHALLGRVVNASTPAEHAKLRRISSAYQVAATLLPIRTVGVQGDCRTYSYAVALSTEKYPPDWKDMQYLAQIIPRICHNVNRVCYAFGGLIKEQVTDITPTFLTQQVVSTLRQADDLATQVLTQSGCAGRIAQMPVVLIPIQFDRDAAVRAASCQRSIVLRPFVTSDFMTGLAALPGSDHMPQDVVDRMCKKLLTVPGISRVLYDLTPKPPATTEWE
Summary
Uniprot
H9JD39
A0A2A4JY22
A0A2A4JZ07
A0A212F8J1
A0A194Q9Y7
A0A2J7QXB7
+ More
A0A2J7QXC1 A0A067RBC0 E0VNC1 A0A0A9X3L7 D6WW83 A0A182IM13 A0A182MZH2 A0A182JWI1 A0A182R7P1 A0A182MLN1 N6U4D8 A0A182YIN0 A0A182TVM7 A0A182XB76 A0A1S4H3V0 A0A1B6HU54 A0A182KJK1 A0A182QQV3 A0A182URM8 A0A1B6J442 A0A1Y1LIU9 A0A182HQW5 A0A182P7T2 A0A0C9RL08 A0A0N7Z908 A0A1W4WUY9 A0A1Q3FWV3 A0A023F412 W5JX82 A0A224XJF3 A0A2M4A214 A0A069DWD7 T1HV64 A0A2M3ZFU4 A0A2M3Z056 A0A2M4A1Y2 A0A2M4BFV5 A0A2M3Z059 A0A1S4J0N8 Q7Q0V2 A0A2M4BGI9 U5EVN1 A0A0V0G619 A0A2A3E812 A0A088ASX2 Q17JX5 A0A182GB75 A0A131XKJ0 A0A0M9ACH3 A0A0J7KS53 A0A1B6CKV9 K7J1R9 A0A1B6CTG3 A0A224YZ73 A0A131YI09 A0A154PF42 A0A026WB09 A0A182FN04 A0A0L7R6G7 L7MGI2 A0A1I8PXJ1 A0A1I8PXP0 A0A3L8DAK4 L7MEX3 A0A0K8TM98 V5GSI7 A0A023GNP4 A0A0P4W848 A0A195CTR3 A0A1I8N4V1 A0A1I8N4U1 T1PP07 A0A151WLS3 A0A023G099 A0A0L0CB16 E2B6F9 F4WJ39 A0A195FPD2 A0A151JR18 A0A1B0A5U4 D3TNY3 A0A158NTU2 A0A1W4UIP0 A0A1L8DXT8 A0A195BMG6 A0A1J1IBR4 B3NKT2 A0A0Q5VXD8 A0A1A9X0R6 Q9VIE7 V5I554 A0A0Q5VU55 A0A1A9Y0D1 A0A1B0AS86
A0A2J7QXC1 A0A067RBC0 E0VNC1 A0A0A9X3L7 D6WW83 A0A182IM13 A0A182MZH2 A0A182JWI1 A0A182R7P1 A0A182MLN1 N6U4D8 A0A182YIN0 A0A182TVM7 A0A182XB76 A0A1S4H3V0 A0A1B6HU54 A0A182KJK1 A0A182QQV3 A0A182URM8 A0A1B6J442 A0A1Y1LIU9 A0A182HQW5 A0A182P7T2 A0A0C9RL08 A0A0N7Z908 A0A1W4WUY9 A0A1Q3FWV3 A0A023F412 W5JX82 A0A224XJF3 A0A2M4A214 A0A069DWD7 T1HV64 A0A2M3ZFU4 A0A2M3Z056 A0A2M4A1Y2 A0A2M4BFV5 A0A2M3Z059 A0A1S4J0N8 Q7Q0V2 A0A2M4BGI9 U5EVN1 A0A0V0G619 A0A2A3E812 A0A088ASX2 Q17JX5 A0A182GB75 A0A131XKJ0 A0A0M9ACH3 A0A0J7KS53 A0A1B6CKV9 K7J1R9 A0A1B6CTG3 A0A224YZ73 A0A131YI09 A0A154PF42 A0A026WB09 A0A182FN04 A0A0L7R6G7 L7MGI2 A0A1I8PXJ1 A0A1I8PXP0 A0A3L8DAK4 L7MEX3 A0A0K8TM98 V5GSI7 A0A023GNP4 A0A0P4W848 A0A195CTR3 A0A1I8N4V1 A0A1I8N4U1 T1PP07 A0A151WLS3 A0A023G099 A0A0L0CB16 E2B6F9 F4WJ39 A0A195FPD2 A0A151JR18 A0A1B0A5U4 D3TNY3 A0A158NTU2 A0A1W4UIP0 A0A1L8DXT8 A0A195BMG6 A0A1J1IBR4 B3NKT2 A0A0Q5VXD8 A0A1A9X0R6 Q9VIE7 V5I554 A0A0Q5VU55 A0A1A9Y0D1 A0A1B0AS86
Pubmed
19121390
22118469
26354079
24845553
20566863
25401762
+ More
18362917 19820115 23537049 25244985 12364791 20966253 28004739 27129103 25474469 20920257 23761445 26334808 17510324 26483478 28049606 20075255 28797301 26830274 24508170 25576852 30249741 26369729 25315136 26108605 20798317 21719571 20353571 21347285 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25765539
18362917 19820115 23537049 25244985 12364791 20966253 28004739 27129103 25474469 20920257 23761445 26334808 17510324 26483478 28049606 20075255 28797301 26830274 24508170 25576852 30249741 26369729 25315136 26108605 20798317 21719571 20353571 21347285 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25765539
EMBL
BABH01027796
BABH01027797
NWSH01000370
PCG76921.1
PCG76918.1
AGBW02009710
+ More
OWR50057.1 KQ459249 KPJ02333.1 NEVH01009382 PNF33227.1 PNF33228.1 KK852618 KDR20132.1 DS235335 EEB14877.1 GBHO01031929 JAG11675.1 KQ971361 EFA09313.1 AXCM01001171 APGK01039913 KB740975 KB632186 ENN76460.1 ERL89746.1 AAAB01008980 GECU01029498 GECU01024747 JAS78208.1 JAS82959.1 AXCN02001996 GECU01013786 JAS93920.1 GEZM01054526 JAV73584.1 APCN01001606 GBYB01008895 JAG78662.1 GDKW01001867 JAI54728.1 GFDL01003113 JAV31932.1 GBBI01002526 JAC16186.1 ADMH02000073 ETN67904.1 GFTR01007704 JAW08722.1 GGFK01001506 MBW34827.1 GBGD01000728 JAC88161.1 ACPB03004450 GGFM01006651 MBW27402.1 GGFM01001159 MBW21910.1 GGFK01001503 MBW34824.1 GGFJ01002794 MBW51935.1 GGFM01001158 MBW21909.1 EAA13830.4 GGFJ01002777 MBW51918.1 GANO01001803 JAB58068.1 GECL01003282 JAP02842.1 KZ288334 PBC27887.1 CH477229 EAT47025.1 JXUM01052185 KQ561723 KXJ77721.1 GEFH01001866 JAP66715.1 KQ435700 KOX80463.1 LBMM01003755 KMQ93173.1 GEDC01023228 JAS14070.1 GEDC01020538 JAS16760.1 GFPF01008128 MAA19274.1 GEDV01009604 JAP78953.1 KQ434890 KZC10469.1 KK107348 EZA52209.1 KQ414646 KOC66419.1 GACK01002656 JAA62378.1 QOIP01000010 RLU17530.1 GACK01002657 JAA62377.1 GDAI01002124 JAI15479.1 GALX01003899 JAB64567.1 GBBM01000600 JAC34818.1 GDRN01074622 JAI63205.1 KQ977279 KYN04083.1 KA649553 KA649728 AFP64357.1 KQ982959 KYQ48770.1 GBBL01000950 JAC26370.1 JRES01000668 KNC29427.1 GL445973 EFN88703.1 GL888181 EGI65809.1 KQ981382 KYN42318.1 KQ978615 KYN29813.1 EZ423135 ADD19411.1 ADTU01026156 ADTU01026157 GFDF01002821 JAV11263.1 KQ976435 KYM87070.1 CVRI01000047 CRK97643.1 CH954179 EDV54455.2 KQS61802.1 AE014134 AY118572 AAF53975.3 AAM49941.1 AHN54634.1 GANP01000660 JAB83808.1 KQS61801.1 JXJN01002692
OWR50057.1 KQ459249 KPJ02333.1 NEVH01009382 PNF33227.1 PNF33228.1 KK852618 KDR20132.1 DS235335 EEB14877.1 GBHO01031929 JAG11675.1 KQ971361 EFA09313.1 AXCM01001171 APGK01039913 KB740975 KB632186 ENN76460.1 ERL89746.1 AAAB01008980 GECU01029498 GECU01024747 JAS78208.1 JAS82959.1 AXCN02001996 GECU01013786 JAS93920.1 GEZM01054526 JAV73584.1 APCN01001606 GBYB01008895 JAG78662.1 GDKW01001867 JAI54728.1 GFDL01003113 JAV31932.1 GBBI01002526 JAC16186.1 ADMH02000073 ETN67904.1 GFTR01007704 JAW08722.1 GGFK01001506 MBW34827.1 GBGD01000728 JAC88161.1 ACPB03004450 GGFM01006651 MBW27402.1 GGFM01001159 MBW21910.1 GGFK01001503 MBW34824.1 GGFJ01002794 MBW51935.1 GGFM01001158 MBW21909.1 EAA13830.4 GGFJ01002777 MBW51918.1 GANO01001803 JAB58068.1 GECL01003282 JAP02842.1 KZ288334 PBC27887.1 CH477229 EAT47025.1 JXUM01052185 KQ561723 KXJ77721.1 GEFH01001866 JAP66715.1 KQ435700 KOX80463.1 LBMM01003755 KMQ93173.1 GEDC01023228 JAS14070.1 GEDC01020538 JAS16760.1 GFPF01008128 MAA19274.1 GEDV01009604 JAP78953.1 KQ434890 KZC10469.1 KK107348 EZA52209.1 KQ414646 KOC66419.1 GACK01002656 JAA62378.1 QOIP01000010 RLU17530.1 GACK01002657 JAA62377.1 GDAI01002124 JAI15479.1 GALX01003899 JAB64567.1 GBBM01000600 JAC34818.1 GDRN01074622 JAI63205.1 KQ977279 KYN04083.1 KA649553 KA649728 AFP64357.1 KQ982959 KYQ48770.1 GBBL01000950 JAC26370.1 JRES01000668 KNC29427.1 GL445973 EFN88703.1 GL888181 EGI65809.1 KQ981382 KYN42318.1 KQ978615 KYN29813.1 EZ423135 ADD19411.1 ADTU01026156 ADTU01026157 GFDF01002821 JAV11263.1 KQ976435 KYM87070.1 CVRI01000047 CRK97643.1 CH954179 EDV54455.2 KQS61802.1 AE014134 AY118572 AAF53975.3 AAM49941.1 AHN54634.1 GANP01000660 JAB83808.1 KQS61801.1 JXJN01002692
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000235965
UP000027135
+ More
UP000009046 UP000007266 UP000075880 UP000075884 UP000075881 UP000075900 UP000075883 UP000019118 UP000030742 UP000076408 UP000075902 UP000076407 UP000075882 UP000075886 UP000075903 UP000075840 UP000075885 UP000192223 UP000000673 UP000015103 UP000007062 UP000242457 UP000005203 UP000008820 UP000069940 UP000249989 UP000053105 UP000036403 UP000002358 UP000076502 UP000053097 UP000069272 UP000053825 UP000095300 UP000279307 UP000078542 UP000095301 UP000075809 UP000037069 UP000008237 UP000007755 UP000078541 UP000078492 UP000092445 UP000005205 UP000192221 UP000078540 UP000183832 UP000008711 UP000091820 UP000000803 UP000092443 UP000092460
UP000009046 UP000007266 UP000075880 UP000075884 UP000075881 UP000075900 UP000075883 UP000019118 UP000030742 UP000076408 UP000075902 UP000076407 UP000075882 UP000075886 UP000075903 UP000075840 UP000075885 UP000192223 UP000000673 UP000015103 UP000007062 UP000242457 UP000005203 UP000008820 UP000069940 UP000249989 UP000053105 UP000036403 UP000002358 UP000076502 UP000053097 UP000069272 UP000053825 UP000095300 UP000279307 UP000078542 UP000095301 UP000075809 UP000037069 UP000008237 UP000007755 UP000078541 UP000078492 UP000092445 UP000005205 UP000192221 UP000078540 UP000183832 UP000008711 UP000091820 UP000000803 UP000092443 UP000092460
PRIDE
Pfam
Interpro
SUPFAM
SSF52317
SSF52317
Gene 3D
ProteinModelPortal
H9JD39
A0A2A4JY22
A0A2A4JZ07
A0A212F8J1
A0A194Q9Y7
A0A2J7QXB7
+ More
A0A2J7QXC1 A0A067RBC0 E0VNC1 A0A0A9X3L7 D6WW83 A0A182IM13 A0A182MZH2 A0A182JWI1 A0A182R7P1 A0A182MLN1 N6U4D8 A0A182YIN0 A0A182TVM7 A0A182XB76 A0A1S4H3V0 A0A1B6HU54 A0A182KJK1 A0A182QQV3 A0A182URM8 A0A1B6J442 A0A1Y1LIU9 A0A182HQW5 A0A182P7T2 A0A0C9RL08 A0A0N7Z908 A0A1W4WUY9 A0A1Q3FWV3 A0A023F412 W5JX82 A0A224XJF3 A0A2M4A214 A0A069DWD7 T1HV64 A0A2M3ZFU4 A0A2M3Z056 A0A2M4A1Y2 A0A2M4BFV5 A0A2M3Z059 A0A1S4J0N8 Q7Q0V2 A0A2M4BGI9 U5EVN1 A0A0V0G619 A0A2A3E812 A0A088ASX2 Q17JX5 A0A182GB75 A0A131XKJ0 A0A0M9ACH3 A0A0J7KS53 A0A1B6CKV9 K7J1R9 A0A1B6CTG3 A0A224YZ73 A0A131YI09 A0A154PF42 A0A026WB09 A0A182FN04 A0A0L7R6G7 L7MGI2 A0A1I8PXJ1 A0A1I8PXP0 A0A3L8DAK4 L7MEX3 A0A0K8TM98 V5GSI7 A0A023GNP4 A0A0P4W848 A0A195CTR3 A0A1I8N4V1 A0A1I8N4U1 T1PP07 A0A151WLS3 A0A023G099 A0A0L0CB16 E2B6F9 F4WJ39 A0A195FPD2 A0A151JR18 A0A1B0A5U4 D3TNY3 A0A158NTU2 A0A1W4UIP0 A0A1L8DXT8 A0A195BMG6 A0A1J1IBR4 B3NKT2 A0A0Q5VXD8 A0A1A9X0R6 Q9VIE7 V5I554 A0A0Q5VU55 A0A1A9Y0D1 A0A1B0AS86
A0A2J7QXC1 A0A067RBC0 E0VNC1 A0A0A9X3L7 D6WW83 A0A182IM13 A0A182MZH2 A0A182JWI1 A0A182R7P1 A0A182MLN1 N6U4D8 A0A182YIN0 A0A182TVM7 A0A182XB76 A0A1S4H3V0 A0A1B6HU54 A0A182KJK1 A0A182QQV3 A0A182URM8 A0A1B6J442 A0A1Y1LIU9 A0A182HQW5 A0A182P7T2 A0A0C9RL08 A0A0N7Z908 A0A1W4WUY9 A0A1Q3FWV3 A0A023F412 W5JX82 A0A224XJF3 A0A2M4A214 A0A069DWD7 T1HV64 A0A2M3ZFU4 A0A2M3Z056 A0A2M4A1Y2 A0A2M4BFV5 A0A2M3Z059 A0A1S4J0N8 Q7Q0V2 A0A2M4BGI9 U5EVN1 A0A0V0G619 A0A2A3E812 A0A088ASX2 Q17JX5 A0A182GB75 A0A131XKJ0 A0A0M9ACH3 A0A0J7KS53 A0A1B6CKV9 K7J1R9 A0A1B6CTG3 A0A224YZ73 A0A131YI09 A0A154PF42 A0A026WB09 A0A182FN04 A0A0L7R6G7 L7MGI2 A0A1I8PXJ1 A0A1I8PXP0 A0A3L8DAK4 L7MEX3 A0A0K8TM98 V5GSI7 A0A023GNP4 A0A0P4W848 A0A195CTR3 A0A1I8N4V1 A0A1I8N4U1 T1PP07 A0A151WLS3 A0A023G099 A0A0L0CB16 E2B6F9 F4WJ39 A0A195FPD2 A0A151JR18 A0A1B0A5U4 D3TNY3 A0A158NTU2 A0A1W4UIP0 A0A1L8DXT8 A0A195BMG6 A0A1J1IBR4 B3NKT2 A0A0Q5VXD8 A0A1A9X0R6 Q9VIE7 V5I554 A0A0Q5VU55 A0A1A9Y0D1 A0A1B0AS86
PDB
2VXO
E-value=0,
Score=2133
Ontologies
PATHWAY
GO
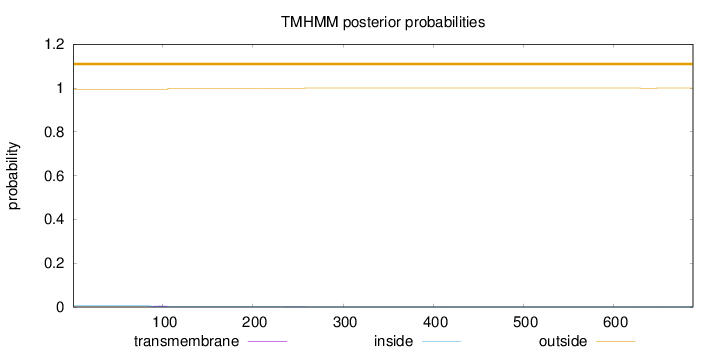
Topology
Length:
688
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10739
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00556
outside
1 - 688
Population Genetic Test Statistics
Pi
196.56374
Theta
177.62456
Tajima's D
0.097715
CLR
16.749748
CSRT
0.391880405979701
Interpretation
Uncertain
