Pre Gene Modal
BGIBMGA007332
Annotation
PREDICTED:_26S_protease_regulatory_subunit_7_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 4.08
Sequence
CDS
ATGCCTGACCATTTAGGGAATGATATGCGCAAAGTGAAAGATGATAAAGAAGAGCCGGAAAAAGAAATAAAATCTCTCGACGAGGGTGATATTGCGCTTTTGAAGTCATACGGCCAAGGACAGTACACAAAAATCATTAAGGAAGTAGAAGAAGGAATTCAGACAGTAATGAAGAGGGTTAATGAACTGACAGGCATCAAAGAGTCAGACACAGGGTTGGCTCCTCCAGCCTTGTGGGATCTAGCAGCTGATAAACAGACACTACAGAATGAACAACCACTACAGGTAGCACGGTGTACGAAAATCATTAATGCAGATTCGAATGATCCAAAATACATAATTAACGTGAAGCAGTTTGCAAAGTTTGTTGTCGATTTACAAGACTCCGTCGCACCCACTGATATTGAAGAAGGCATGAGGGTTGGTGTGGATCGTAATAAATACCAGATCCACATACCCCTGCCTCCGAAAATAGATCCAACGGTCACCATGATGCAGGTCGAGGAGAAACCTGATGTAACATACAGTGACGTGGGTGGATGTAAAGAGCAGATAGAAAAACTAAGAGAAGTTGTTGAGACACCACTGTTGCATCCGGAGAAATTCGTGAAGCTGGGTATCGAGCCCCCGAAGGGAGTGTTGCTATTCGGCCCCCCCGGGACCGGGAAGACGCTGTGTGCTCGCGCCGTCGCCAACCGGACGGACGCCTGCTTCATTCGGGTTATTGGTTCTGAATTAGTGCAGAAATATGTCGGCGAAGGGGCCCGGATGGTGCGAGAACTGTTCGAAATGGCTCGTAGCAAGAAGGCCTGTCTTATATTCTTCGATGAAATCGACGCTATAGGTGGCGCCAGGTTCGACGATGGTGCTGGCGGCGATAATGAAGTTCAGAGAACGATGTTGGAGCTGATCAACCAGTTGGACGGCTTCGATCCTCGCGGCAATATTAAGGTGCTGATGGCCACGAACCGTCCCGACACGCTGGACCCGGCGCTGATGCGGCCGGGGCGGCTGGACCGCAAGGTGGAGTTCGGGCTGCCGGACCTGGAGGGGCGCGCCCACATCTTCCGCATCCACGCGCGCTCCATGAGCGTCGAGCGAGACATCCGGTTCGACCTGCTCGCCAGGCTCTGTCCCAACTCCACCGGCGCCGAGATCAGGTCTGTTTGCACGGAGGCGGGCATGTTCGCGATCCGCGCCCGCCGCAAGGTTGCCACCGAGAAGGACTTCCTCGAAGCCGTCAACAAAGTCATCAAGTCGTACGCCAAGTTCTCAGCGACACCCAGATACATGACCTACAACTAA
Protein
MPDHLGNDMRKVKDDKEEPEKEIKSLDEGDIALLKSYGQGQYTKIIKEVEEGIQTVMKRVNELTGIKESDTGLAPPALWDLAADKQTLQNEQPLQVARCTKIINADSNDPKYIINVKQFAKFVVDLQDSVAPTDIEEGMRVGVDRNKYQIHIPLPPKIDPTVTMMQVEEKPDVTYSDVGGCKEQIEKLREVVETPLLHPEKFVKLGIEPPKGVLLFGPPGTGKTLCARAVANRTDACFIRVIGSELVQKYVGEGARMVRELFEMARSKKACLIFFDEIDAIGGARFDDGAGGDNEVQRTMLELINQLDGFDPRGNIKVLMATNRPDTLDPALMRPGRLDRKVEFGLPDLEGRAHIFRIHARSMSVERDIRFDLLARLCPNSTGAEIRSVCTEAGMFAIRARRKVATEKDFLEAVNKVIKSYAKFSATPRYMTYN
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
H9JCT5
A0A2W1BK31
I4DK92
A0A2A4JY24
A0A212F8L2
S4PLZ6
+ More
D6WX64 N6T8Q7 A0A2A3E592 A0A088A957 F4X0C5 A0A0J7L469 A0A1B6FD73 A0A158NFM1 A0A1B6IAH1 E9IEV2 A0A2J7PIB4 E2C0K3 A0A026WS10 A0A067R996 A0A154PI70 A0A1W4W4H6 A0A1B6KEK1 E2AS83 A0A1B6DYY9 A0A195AXX0 A0A1Y1M958 A0A151IY01 A0A151JWT7 A0A151WW00 A0A0L7QPZ4 A0A0P4W2J9 A0A023FCX8 A0A069DYZ4 A0A0C9PZB6 A0A182JNJ2 A0A336N327 A0A0A9Y5F1 A0A310SBC5 A0A195D876 A0A182J2Q3 E9FY58 A0A0P5M1A9 A0A182MCK7 A0A182RTE5 A0A182V785 A0A182YB38 A0A182WDH3 A0A182HWE0 A0A182SCZ8 A0A182NZK7 A0A182U702 A0A182KL62 A0A182X5K9 Q7QHG0 A0A232EY04 A0A182N2D1 A0A182Q4L4 K7J0F0 A0A0P5NYS3 A0A0N1PI44 A0A1I9WLG6 U4UER6 A0A1L8DSU0 Q16KL0 A0A023ES69 R4WRD0 U5EX53 A0A0N8DN17 A0A2M3Z649 A0A2M4AF89 A0A2M4BNG1 W5J9A8 A0A182FKJ2 A0A1J1IF22 A0A2P6KE78 G3MH24 A0A0Q9XH06 R4G5R5 A0A131XJW4 A0A023GN27 G3MM95 A0A224YLS1 A0A131Z248 L7M8R4 T1HS78 A0A023FWB2 A0A1Q3FED3 A0A0K8TN06 B0X195 A0A0A1XS41 A0A210Q5J9 W8BU79 A0A034V3F2 A0A0C9RBT8 A0A0L0BKI7 A0A0P4VYN7 A0A0M9A0A5 A0A0N8CT62 A0A0K8WBX7
D6WX64 N6T8Q7 A0A2A3E592 A0A088A957 F4X0C5 A0A0J7L469 A0A1B6FD73 A0A158NFM1 A0A1B6IAH1 E9IEV2 A0A2J7PIB4 E2C0K3 A0A026WS10 A0A067R996 A0A154PI70 A0A1W4W4H6 A0A1B6KEK1 E2AS83 A0A1B6DYY9 A0A195AXX0 A0A1Y1M958 A0A151IY01 A0A151JWT7 A0A151WW00 A0A0L7QPZ4 A0A0P4W2J9 A0A023FCX8 A0A069DYZ4 A0A0C9PZB6 A0A182JNJ2 A0A336N327 A0A0A9Y5F1 A0A310SBC5 A0A195D876 A0A182J2Q3 E9FY58 A0A0P5M1A9 A0A182MCK7 A0A182RTE5 A0A182V785 A0A182YB38 A0A182WDH3 A0A182HWE0 A0A182SCZ8 A0A182NZK7 A0A182U702 A0A182KL62 A0A182X5K9 Q7QHG0 A0A232EY04 A0A182N2D1 A0A182Q4L4 K7J0F0 A0A0P5NYS3 A0A0N1PI44 A0A1I9WLG6 U4UER6 A0A1L8DSU0 Q16KL0 A0A023ES69 R4WRD0 U5EX53 A0A0N8DN17 A0A2M3Z649 A0A2M4AF89 A0A2M4BNG1 W5J9A8 A0A182FKJ2 A0A1J1IF22 A0A2P6KE78 G3MH24 A0A0Q9XH06 R4G5R5 A0A131XJW4 A0A023GN27 G3MM95 A0A224YLS1 A0A131Z248 L7M8R4 T1HS78 A0A023FWB2 A0A1Q3FED3 A0A0K8TN06 B0X195 A0A0A1XS41 A0A210Q5J9 W8BU79 A0A034V3F2 A0A0C9RBT8 A0A0L0BKI7 A0A0P4VYN7 A0A0M9A0A5 A0A0N8CT62 A0A0K8WBX7
Pubmed
19121390
28756777
22651552
26354079
22118469
23622113
+ More
18362917 19820115 23537049 21719571 21347285 21282665 20798317 24508170 24845553 28004739 27129103 25474469 26334808 25401762 26823975 21292972 25244985 20966253 12364791 14747013 17210077 28648823 20075255 27538518 17510324 24945155 26483478 23691247 20920257 23761445 22216098 17994087 28049606 28797301 26830274 25576852 26369729 25830018 28812685 24495485 25348373 26108605
18362917 19820115 23537049 21719571 21347285 21282665 20798317 24508170 24845553 28004739 27129103 25474469 26334808 25401762 26823975 21292972 25244985 20966253 12364791 14747013 17210077 28648823 20075255 27538518 17510324 24945155 26483478 23691247 20920257 23761445 22216098 17994087 28049606 28797301 26830274 25576852 26369729 25830018 28812685 24495485 25348373 26108605
EMBL
BABH01027796
KZ150074
PZC74004.1
AK401710
KQ459249
BAM18332.1
+ More
KPJ02332.1 NWSH01000370 PCG76917.1 AGBW02009710 OWR50058.1 GAIX01003795 JAA88765.1 KQ971361 EFA08798.1 APGK01039579 KB740972 ENN76609.1 KZ288402 PBC26241.1 GL888497 EGI60083.1 LBMM01000707 KMQ97687.1 GECZ01021705 JAS48064.1 ADTU01014663 GECU01023787 JAS83919.1 GL762722 EFZ20880.1 NEVH01025128 PNF16072.1 GL451800 EFN78520.1 KK107119 EZA58812.1 KK852618 KDR20131.1 KQ434924 KZC11539.1 GEBQ01030118 JAT09859.1 GL442229 EFN63713.1 GEDC01006398 JAS30900.1 KQ976716 KYM76885.1 GEZM01042422 JAV79867.1 KQ980784 KYN13176.1 KQ981620 KYN39220.1 KQ982699 KYQ51947.1 KQ414796 KOC60708.1 GDKW01000856 JAI55739.1 GBBI01000178 JAC18534.1 GBGD01001535 JAC87354.1 GBYB01006873 JAG76640.1 UFQT01000644 UFQT01004045 SSX26047.1 SSX35453.1 GBHO01016743 GBRD01016557 GDHC01013392 JAG26861.1 JAG49269.1 JAQ05237.1 KQ761824 OAD56755.1 KQ976750 KYN08649.1 GL732527 EFX87834.1 GDIQ01186466 GDIQ01106692 LRGB01002371 JAK65259.1 KZS08056.1 AXCM01000599 APCN01001485 AAAB01008816 EAA05145.2 NNAY01001637 OXU23354.1 AXCN02001727 GDIQ01135940 JAL15786.1 KQ460468 KPJ14591.1 KU932350 APA33986.1 KB632061 ERL88395.1 GFDF01004556 JAV09528.1 CH477951 EAT34852.1 JXUM01135370 GAPW01001526 KQ568261 JAC12072.1 KXJ69072.1 AK417232 BAN20447.1 GANO01001110 JAB58761.1 GDIP01017711 JAM86004.1 GGFM01003242 MBW23993.1 GGFK01006138 MBW39459.1 GGFJ01005476 MBW54617.1 ADMH02002090 ETN59374.1 CVRI01000047 CRK97598.1 MWRG01014509 PRD24607.1 JO841175 AEO32792.1 CH933808 KRG04500.1 GAHY01000300 JAA77210.1 GEFH01001929 JAP66652.1 GBBM01001148 JAC34270.1 JO842996 AEO34613.1 GFPF01005543 MAA16689.1 GEDV01003737 JAP84820.1 GACK01004782 JAA60252.1 ACPB03021813 GBBL01001169 JAC26151.1 GFDL01009128 JAV25917.1 GDAI01001869 JAI15734.1 DS232256 EDS38544.1 GBXI01000944 JAD13348.1 NEDP02004925 OWF44010.1 GAMC01001635 JAC04921.1 GAKP01022677 JAC36275.1 GBYB01014039 JAG83806.1 JRES01001712 KNC20567.1 GDRN01102684 JAI58201.1 KQ435802 KOX73209.1 GDIP01101351 JAM02364.1 GDHF01014135 GDHF01003621 JAI38179.1 JAI48693.1
KPJ02332.1 NWSH01000370 PCG76917.1 AGBW02009710 OWR50058.1 GAIX01003795 JAA88765.1 KQ971361 EFA08798.1 APGK01039579 KB740972 ENN76609.1 KZ288402 PBC26241.1 GL888497 EGI60083.1 LBMM01000707 KMQ97687.1 GECZ01021705 JAS48064.1 ADTU01014663 GECU01023787 JAS83919.1 GL762722 EFZ20880.1 NEVH01025128 PNF16072.1 GL451800 EFN78520.1 KK107119 EZA58812.1 KK852618 KDR20131.1 KQ434924 KZC11539.1 GEBQ01030118 JAT09859.1 GL442229 EFN63713.1 GEDC01006398 JAS30900.1 KQ976716 KYM76885.1 GEZM01042422 JAV79867.1 KQ980784 KYN13176.1 KQ981620 KYN39220.1 KQ982699 KYQ51947.1 KQ414796 KOC60708.1 GDKW01000856 JAI55739.1 GBBI01000178 JAC18534.1 GBGD01001535 JAC87354.1 GBYB01006873 JAG76640.1 UFQT01000644 UFQT01004045 SSX26047.1 SSX35453.1 GBHO01016743 GBRD01016557 GDHC01013392 JAG26861.1 JAG49269.1 JAQ05237.1 KQ761824 OAD56755.1 KQ976750 KYN08649.1 GL732527 EFX87834.1 GDIQ01186466 GDIQ01106692 LRGB01002371 JAK65259.1 KZS08056.1 AXCM01000599 APCN01001485 AAAB01008816 EAA05145.2 NNAY01001637 OXU23354.1 AXCN02001727 GDIQ01135940 JAL15786.1 KQ460468 KPJ14591.1 KU932350 APA33986.1 KB632061 ERL88395.1 GFDF01004556 JAV09528.1 CH477951 EAT34852.1 JXUM01135370 GAPW01001526 KQ568261 JAC12072.1 KXJ69072.1 AK417232 BAN20447.1 GANO01001110 JAB58761.1 GDIP01017711 JAM86004.1 GGFM01003242 MBW23993.1 GGFK01006138 MBW39459.1 GGFJ01005476 MBW54617.1 ADMH02002090 ETN59374.1 CVRI01000047 CRK97598.1 MWRG01014509 PRD24607.1 JO841175 AEO32792.1 CH933808 KRG04500.1 GAHY01000300 JAA77210.1 GEFH01001929 JAP66652.1 GBBM01001148 JAC34270.1 JO842996 AEO34613.1 GFPF01005543 MAA16689.1 GEDV01003737 JAP84820.1 GACK01004782 JAA60252.1 ACPB03021813 GBBL01001169 JAC26151.1 GFDL01009128 JAV25917.1 GDAI01001869 JAI15734.1 DS232256 EDS38544.1 GBXI01000944 JAD13348.1 NEDP02004925 OWF44010.1 GAMC01001635 JAC04921.1 GAKP01022677 JAC36275.1 GBYB01014039 JAG83806.1 JRES01001712 KNC20567.1 GDRN01102684 JAI58201.1 KQ435802 KOX73209.1 GDIP01101351 JAM02364.1 GDHF01014135 GDHF01003621 JAI38179.1 JAI48693.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000007266
UP000019118
+ More
UP000242457 UP000005203 UP000007755 UP000036403 UP000005205 UP000235965 UP000008237 UP000053097 UP000027135 UP000076502 UP000192223 UP000000311 UP000078540 UP000078492 UP000078541 UP000075809 UP000053825 UP000075881 UP000078542 UP000075880 UP000000305 UP000076858 UP000075883 UP000075900 UP000075903 UP000076408 UP000075920 UP000075840 UP000075901 UP000075885 UP000075902 UP000075882 UP000076407 UP000007062 UP000215335 UP000075884 UP000075886 UP000002358 UP000053240 UP000030742 UP000008820 UP000069940 UP000249989 UP000000673 UP000069272 UP000183832 UP000009192 UP000015103 UP000002320 UP000242188 UP000037069 UP000053105
UP000242457 UP000005203 UP000007755 UP000036403 UP000005205 UP000235965 UP000008237 UP000053097 UP000027135 UP000076502 UP000192223 UP000000311 UP000078540 UP000078492 UP000078541 UP000075809 UP000053825 UP000075881 UP000078542 UP000075880 UP000000305 UP000076858 UP000075883 UP000075900 UP000075903 UP000076408 UP000075920 UP000075840 UP000075901 UP000075885 UP000075902 UP000075882 UP000076407 UP000007062 UP000215335 UP000075884 UP000075886 UP000002358 UP000053240 UP000030742 UP000008820 UP000069940 UP000249989 UP000000673 UP000069272 UP000183832 UP000009192 UP000015103 UP000002320 UP000242188 UP000037069 UP000053105
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9JCT5
A0A2W1BK31
I4DK92
A0A2A4JY24
A0A212F8L2
S4PLZ6
+ More
D6WX64 N6T8Q7 A0A2A3E592 A0A088A957 F4X0C5 A0A0J7L469 A0A1B6FD73 A0A158NFM1 A0A1B6IAH1 E9IEV2 A0A2J7PIB4 E2C0K3 A0A026WS10 A0A067R996 A0A154PI70 A0A1W4W4H6 A0A1B6KEK1 E2AS83 A0A1B6DYY9 A0A195AXX0 A0A1Y1M958 A0A151IY01 A0A151JWT7 A0A151WW00 A0A0L7QPZ4 A0A0P4W2J9 A0A023FCX8 A0A069DYZ4 A0A0C9PZB6 A0A182JNJ2 A0A336N327 A0A0A9Y5F1 A0A310SBC5 A0A195D876 A0A182J2Q3 E9FY58 A0A0P5M1A9 A0A182MCK7 A0A182RTE5 A0A182V785 A0A182YB38 A0A182WDH3 A0A182HWE0 A0A182SCZ8 A0A182NZK7 A0A182U702 A0A182KL62 A0A182X5K9 Q7QHG0 A0A232EY04 A0A182N2D1 A0A182Q4L4 K7J0F0 A0A0P5NYS3 A0A0N1PI44 A0A1I9WLG6 U4UER6 A0A1L8DSU0 Q16KL0 A0A023ES69 R4WRD0 U5EX53 A0A0N8DN17 A0A2M3Z649 A0A2M4AF89 A0A2M4BNG1 W5J9A8 A0A182FKJ2 A0A1J1IF22 A0A2P6KE78 G3MH24 A0A0Q9XH06 R4G5R5 A0A131XJW4 A0A023GN27 G3MM95 A0A224YLS1 A0A131Z248 L7M8R4 T1HS78 A0A023FWB2 A0A1Q3FED3 A0A0K8TN06 B0X195 A0A0A1XS41 A0A210Q5J9 W8BU79 A0A034V3F2 A0A0C9RBT8 A0A0L0BKI7 A0A0P4VYN7 A0A0M9A0A5 A0A0N8CT62 A0A0K8WBX7
D6WX64 N6T8Q7 A0A2A3E592 A0A088A957 F4X0C5 A0A0J7L469 A0A1B6FD73 A0A158NFM1 A0A1B6IAH1 E9IEV2 A0A2J7PIB4 E2C0K3 A0A026WS10 A0A067R996 A0A154PI70 A0A1W4W4H6 A0A1B6KEK1 E2AS83 A0A1B6DYY9 A0A195AXX0 A0A1Y1M958 A0A151IY01 A0A151JWT7 A0A151WW00 A0A0L7QPZ4 A0A0P4W2J9 A0A023FCX8 A0A069DYZ4 A0A0C9PZB6 A0A182JNJ2 A0A336N327 A0A0A9Y5F1 A0A310SBC5 A0A195D876 A0A182J2Q3 E9FY58 A0A0P5M1A9 A0A182MCK7 A0A182RTE5 A0A182V785 A0A182YB38 A0A182WDH3 A0A182HWE0 A0A182SCZ8 A0A182NZK7 A0A182U702 A0A182KL62 A0A182X5K9 Q7QHG0 A0A232EY04 A0A182N2D1 A0A182Q4L4 K7J0F0 A0A0P5NYS3 A0A0N1PI44 A0A1I9WLG6 U4UER6 A0A1L8DSU0 Q16KL0 A0A023ES69 R4WRD0 U5EX53 A0A0N8DN17 A0A2M3Z649 A0A2M4AF89 A0A2M4BNG1 W5J9A8 A0A182FKJ2 A0A1J1IF22 A0A2P6KE78 G3MH24 A0A0Q9XH06 R4G5R5 A0A131XJW4 A0A023GN27 G3MM95 A0A224YLS1 A0A131Z248 L7M8R4 T1HS78 A0A023FWB2 A0A1Q3FED3 A0A0K8TN06 B0X195 A0A0A1XS41 A0A210Q5J9 W8BU79 A0A034V3F2 A0A0C9RBT8 A0A0L0BKI7 A0A0P4VYN7 A0A0M9A0A5 A0A0N8CT62 A0A0K8WBX7
PDB
6MSK
E-value=0,
Score=1837
Ontologies
KEGG
GO
PANTHER
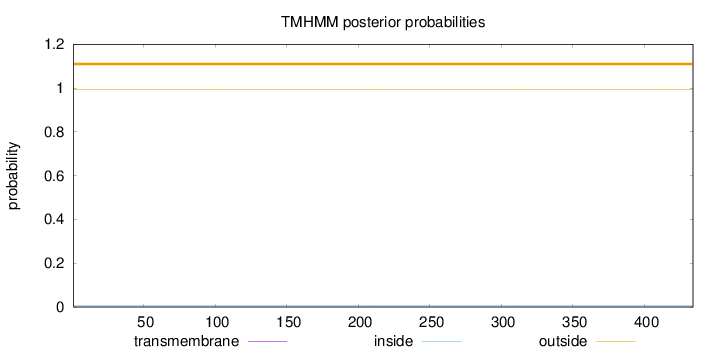
Topology
Length:
434
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00129
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00621
outside
1 - 434
Population Genetic Test Statistics
Pi
243.044731
Theta
162.100226
Tajima's D
1.564692
CLR
0.573495
CSRT
0.805009749512524
Interpretation
Uncertain
