Gene
KWMTBOMO01110
Pre Gene Modal
BGIBMGA007335
Annotation
PREDICTED:_espin_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.399
Sequence
CDS
ATGGATAATGACGTGACGCCGGTATACCTGGCCGCGCAGGAAGGTCATCTTGCAGTGCTGAAGTACTTGGTCTTGGAGGCCGGGGGGCGATTGGACGCGAGAGCCAGAGATGGCATGCTTCCGATACACGCTGCCGCCCAGACCGGCTGCCTCGACTGTGTCAAATGGATGATTATAGAGCGCGGCGTTGACCCGAACGCGAGGGATGGAGACGGTGCGACTCCTCTTCATTTCGCAGCTTCGAGGGGTCATCTCAGTACCGTTCGCTGGTTACTGAGACACGGAGCCAGACTCCATCTGGACAGGCACGGCAAGAGCCCTATTAATGATGCCGCAGAAAATCATCACTTGGAATGCCTGAACGTGCTGGTCGCGGCTGGCGCCGCCGGAGCTGACAGCAAACAGCTCTCCTCGTATAAATCCATCCACTCCTGCGCTTGCACGCCTGGGGATGCGCGGTGCGCGTTACCGAACTGCGTTAACGCGATCAGCACTCGGTCCCCATTTTACCTCCACGGCCCGGAGAGGGAGCGCCCCGCGACCCAGGACAGGCGGGGGTCTCTACCCTCCACGGGTGGCTCCGTTAGCTCAGCGCGCTCCGGGCCCGCCGACGGACTCTACGTCAACCCCATGCAACGAGCCACTTCTACCAACGTCGTACACCATACGTCTTCCGGAGAAGATAGTTCAGGCTGTTCAGCATCCGAGGCAGACACGGCTAACTCCGGAGGCTGGTTCCTTCACGGCAGCAACAACGGTAGCACCGGTGCTCCTGCCGCTTTGTACCAGCGCGTCAGAGAGCTGTTTCATTCGCGCTCTCCTGGCCCGAGGGTCCCGGCTGAAGGACAGGAAGCTCCCACGATGACAGTTAAAGCAGAAGTGCACGGCACGGCAGAAGTGACAACGGTCCAAGAACAATCCGACAGCGACAGTGGCGCTGAAGCGAATAAGAGGGATCATCATCATTATGAAGATATTTACCTGCCTCGAGAAGAGAATCAGAGGAGAAGGAGTAGTTCTCGGGACTCTGGAAGTCACAGTCGCCCCGGCAGCGCGGCTCTCCTCGTGGACTACGACTCCAAAGATAGCAACGACAGGTACGGGATATTATTATTATGTTAA
Protein
MDNDVTPVYLAAQEGHLAVLKYLVLEAGGRLDARARDGMLPIHAAAQTGCLDCVKWMIIERGVDPNARDGDGATPLHFAASRGHLSTVRWLLRHGARLHLDRHGKSPINDAAENHHLECLNVLVAAGAAGADSKQLSSYKSIHSCACTPGDARCALPNCVNAISTRSPFYLHGPERERPATQDRRGSLPSTGGSVSSARSGPADGLYVNPMQRATSTNVVHHTSSGEDSSGCSASEADTANSGGWFLHGSNNGSTGAPAALYQRVRELFHSRSPGPRVPAEGQEAPTMTVKAEVHGTAEVTTVQEQSDSDSGAEANKRDHHHYEDIYLPREENQRRRSSSRDSGSHSRPGSAALLVDYDSKDSNDRYGILLLC
Summary
Uniprot
A0A2H1WAJ7
H9JCT8
A0A2A4JG88
A0A194R502
N6TMJ8
A0A139WD96
+ More
T1GUT6 A0A139WDK3 U4UBY2 A0A1Y1M366 A0A1Y1M816 A0A1Y1M147 A0A182GQ86 A0A067R912 A0A1S4G6Z1 A0A1B0GQJ2 A0A2J7QB45 A0A2J7QB44 A0A1B0CC07 A0A034W0L8 A0A034W180 A0A0K8WMP6 A0A0K8UG53 A0A0K8V922 A0A0K8VPS4 A0A0K8VDE1 A0A0K8VQF5 A0A2P8Z5K0 Q177D8 A0A336LVC9 A0A182FMJ5 A0A1J1ILQ5 A0A182MGY5 W8BG48 W8C2F8 W8BB56 A0A0A1X620 A0A0A1WY35 A0A182RVW2
T1GUT6 A0A139WDK3 U4UBY2 A0A1Y1M366 A0A1Y1M816 A0A1Y1M147 A0A182GQ86 A0A067R912 A0A1S4G6Z1 A0A1B0GQJ2 A0A2J7QB45 A0A2J7QB44 A0A1B0CC07 A0A034W0L8 A0A034W180 A0A0K8WMP6 A0A0K8UG53 A0A0K8V922 A0A0K8VPS4 A0A0K8VDE1 A0A0K8VQF5 A0A2P8Z5K0 Q177D8 A0A336LVC9 A0A182FMJ5 A0A1J1ILQ5 A0A182MGY5 W8BG48 W8C2F8 W8BB56 A0A0A1X620 A0A0A1WY35 A0A182RVW2
Pubmed
EMBL
ODYU01007070
SOQ49514.1
BABH01027779
BABH01027780
NWSH01001582
PCG70796.1
+ More
KQ460685 KPJ12883.1 APGK01018213 KB740071 ENN81709.1 KQ971361 KYB25920.1 CAQQ02098507 CAQQ02098508 KYB25921.1 KB632286 ERL91434.1 GEZM01042946 JAV79458.1 GEZM01042947 JAV79457.1 GEZM01042945 JAV79459.1 JXUM01080314 JXUM01080315 JXUM01080316 JXUM01080317 KQ563178 KXJ74330.1 KK852618 KDR20150.1 AJVK01007288 AJVK01007289 AJVK01007290 AJVK01007291 NEVH01016302 PNF25806.1 PNF25803.1 AJWK01006031 AJWK01006032 GAKP01011070 JAC47882.1 GAKP01011067 JAC47885.1 GDHF01000059 JAI52255.1 GDHF01031370 GDHF01029078 GDHF01027034 GDHF01014401 GDHF01004568 JAI20944.1 JAI23236.1 JAI25280.1 JAI37913.1 JAI47746.1 GDHF01016948 JAI35366.1 GDHF01027713 GDHF01011442 JAI24601.1 JAI40872.1 GDHF01015462 JAI36852.1 GDHF01011534 JAI40780.1 PYGN01000186 PSN51771.1 CH477377 EAT42287.1 UFQT01000154 SSX20961.1 CVRI01000054 CRL00006.1 AXCM01010818 GAMC01010632 JAB95923.1 GAMC01010631 JAB95924.1 GAMC01010633 JAB95922.1 GBXI01008006 JAD06286.1 GBXI01010872 JAD03420.1
KQ460685 KPJ12883.1 APGK01018213 KB740071 ENN81709.1 KQ971361 KYB25920.1 CAQQ02098507 CAQQ02098508 KYB25921.1 KB632286 ERL91434.1 GEZM01042946 JAV79458.1 GEZM01042947 JAV79457.1 GEZM01042945 JAV79459.1 JXUM01080314 JXUM01080315 JXUM01080316 JXUM01080317 KQ563178 KXJ74330.1 KK852618 KDR20150.1 AJVK01007288 AJVK01007289 AJVK01007290 AJVK01007291 NEVH01016302 PNF25806.1 PNF25803.1 AJWK01006031 AJWK01006032 GAKP01011070 JAC47882.1 GAKP01011067 JAC47885.1 GDHF01000059 JAI52255.1 GDHF01031370 GDHF01029078 GDHF01027034 GDHF01014401 GDHF01004568 JAI20944.1 JAI23236.1 JAI25280.1 JAI37913.1 JAI47746.1 GDHF01016948 JAI35366.1 GDHF01027713 GDHF01011442 JAI24601.1 JAI40872.1 GDHF01015462 JAI36852.1 GDHF01011534 JAI40780.1 PYGN01000186 PSN51771.1 CH477377 EAT42287.1 UFQT01000154 SSX20961.1 CVRI01000054 CRL00006.1 AXCM01010818 GAMC01010632 JAB95923.1 GAMC01010631 JAB95924.1 GAMC01010633 JAB95922.1 GBXI01008006 JAD06286.1 GBXI01010872 JAD03420.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
A0A2H1WAJ7
H9JCT8
A0A2A4JG88
A0A194R502
N6TMJ8
A0A139WD96
+ More
T1GUT6 A0A139WDK3 U4UBY2 A0A1Y1M366 A0A1Y1M816 A0A1Y1M147 A0A182GQ86 A0A067R912 A0A1S4G6Z1 A0A1B0GQJ2 A0A2J7QB45 A0A2J7QB44 A0A1B0CC07 A0A034W0L8 A0A034W180 A0A0K8WMP6 A0A0K8UG53 A0A0K8V922 A0A0K8VPS4 A0A0K8VDE1 A0A0K8VQF5 A0A2P8Z5K0 Q177D8 A0A336LVC9 A0A182FMJ5 A0A1J1ILQ5 A0A182MGY5 W8BG48 W8C2F8 W8BB56 A0A0A1X620 A0A0A1WY35 A0A182RVW2
T1GUT6 A0A139WDK3 U4UBY2 A0A1Y1M366 A0A1Y1M816 A0A1Y1M147 A0A182GQ86 A0A067R912 A0A1S4G6Z1 A0A1B0GQJ2 A0A2J7QB45 A0A2J7QB44 A0A1B0CC07 A0A034W0L8 A0A034W180 A0A0K8WMP6 A0A0K8UG53 A0A0K8V922 A0A0K8VPS4 A0A0K8VDE1 A0A0K8VQF5 A0A2P8Z5K0 Q177D8 A0A336LVC9 A0A182FMJ5 A0A1J1ILQ5 A0A182MGY5 W8BG48 W8C2F8 W8BB56 A0A0A1X620 A0A0A1WY35 A0A182RVW2
PDB
5ET1
E-value=3.26141e-26,
Score=294
Ontologies
GO
Topology
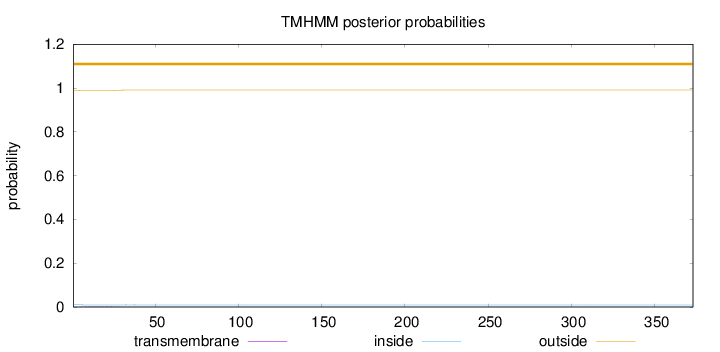
Length:
373
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0336900000000002
Exp number, first 60 AAs:
0.03315
Total prob of N-in:
0.01034
outside
1 - 373
Population Genetic Test Statistics
Pi
233.302517
Theta
168.556129
Tajima's D
1.348076
CLR
0.087442
CSRT
0.753612319384031
Interpretation
Uncertain
