Gene
KWMTBOMO01106
Pre Gene Modal
BGIBMGA007431
Annotation
PREDICTED:_aladin_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.355 PlasmaMembrane Reliability : 1.794
Sequence
CDS
ATGAACTTCCCAAACCTACCTGGTCCAGAAGAAACTGCTTTTTGTAAATTAAATGAAGTATATTGTTGCGGCAATGCCCGATATGGTAATATTTCAACATTTTCAAATGCGACGAAAAAACATCCGATAATCAACGTGACTAGAGACTTACATCATCACAGAACATCAGACGAGAATATTTCCATGTACATAGATGTAGAAGATAATTTGCTCAAAAAAATTACTAGTGTTTGGTACAAACAAGGATTTTTAGAAGCTCTAAGTGTAGCCGCCGACCCTTCTGTTAATAGAGAAAGTCCAACATTGGCAGTCACAGCATCTTACATCTTAAAAGTAGCAAATATTTTAACCGCCTTCAGATATTTCTTACAGCCCCATTTAAAAGATATAGGTCCTAAAATTGTCGCAAATTATTCAAGAACTAGGAACTGGGGTCAAGTGCCAGTCAAATGCTTAGCATGGCATCCTCACACAACAAAAATTGCTGTAGCGACTGTCGATGATAATGTGAGAGTGTATTGTTCAGAAGTAACATTTGTTTCTACATTGAAGTGTAAAGCACAAGGTCATGTGTCATCACTGAACTGGAGACCTCTATCTGCTTCTGAGCTGGCTGTAGGTTGTGAGCAAGGAGTCATTGTATGGACCGTCGACCCAAATTTGATGTTCACAAAACCTTCATCAAGTAATGCTGTTCTATTGAAAAAAAATGGGCACAGTCCAGTAACAGACGTGAGCTGGTCTCCAAACGGTGACCTCTTGATAAGTTGTAGTGGTGCTGACACTTCCATGCTAGTTTGGGATACAGCAATGGAAACAGCCATACCACTGCGGAGAGTTGCTGGTGGAGGCAACGTGTTTGCCAGATGGTCTTTTGATGCTAGCAAAATCTTCACTGCAACATCTTCAATTATATTTCGAATTTGGGACACAAAAACCTGGACTCCGGAAAGATGGTGCGCTCGTGGACATCGAGTTGTAGCTGCTTGTTGGGGACCTGACAATATCTTGCTGTTTGCTGCCAAAGGTGAACCAATGGTTTACGCCCTCACAACCACAAGTCTTCTGAGTGGATCCCAAACTACGAAAGCTACACCAGCATTAGACGTGACAAAGGTGGAACTGTCATCAGGAGAGCCAGTCGGTGGACCCATATTGGATATGTGTTGGGATCCTACCGGCCGTTACATGGCGATGCTGTTTGAAGAATCACATCTCGTTGCTGTCTTCTGTACCACATATATCATGATGCAACTGAAAATTGATCCATGCTGTTTCGTGACTGGCGTAGAAGACGAAATACCCTGTACAATGGCCTTCCAGCAAAACTTCAACGACGGAGCGTGTCTCACGATCGCCTGGTCCTCCGGCAGAGTGCAACACTTTCCCATCATATACTCCGAGTCGTACTGA
Protein
MNFPNLPGPEETAFCKLNEVYCCGNARYGNISTFSNATKKHPIINVTRDLHHHRTSDENISMYIDVEDNLLKKITSVWYKQGFLEALSVAADPSVNRESPTLAVTASYILKVANILTAFRYFLQPHLKDIGPKIVANYSRTRNWGQVPVKCLAWHPHTTKIAVATVDDNVRVYCSEVTFVSTLKCKAQGHVSSLNWRPLSASELAVGCEQGVIVWTVDPNLMFTKPSSSNAVLLKKNGHSPVTDVSWSPNGDLLISCSGADTSMLVWDTAMETAIPLRRVAGGGNVFARWSFDASKIFTATSSIIFRIWDTKTWTPERWCARGHRVVAACWGPDNILLFAAKGEPMVYALTTTSLLSGSQTTKATPALDVTKVELSSGEPVGGPILDMCWDPTGRYMAMLFEESHLVAVFCTTYIMMQLKIDPCCFVTGVEDEIPCTMAFQQNFNDGACLTIAWSSGRVQHFPIIYSESY
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
H9JD34
A0A2A4J051
A0A2W1BK21
A0A212FBE0
A0A0L7KNH6
S4PX74
+ More
A0A0N0P9W5 A0A194R5U0 A0A2H1VBA6 A0A067R739 E0VE24 A0A1B6C7V6 A0A1B6FTZ0 A0A1B6IPA1 A0A1B6LSC7 D6WNN3 A0A1Y1ND83 A0A0C9RF90 A0A1W4XLM8 A0A1S4F6T6 Q17DT7 A0A154PK45 A0A182GNX3 A0A182GCA7 A0A0L7R2H4 A0A087ZNG8 A0A1Q3EWE5 A0A0V0GAL8 B0XBT7 A0A069DU43 K7INI6 W8CBC7 A0A023EZB0 A0A1S3HFA3 A0A232FBH5 A0A034UZI2 A0A2A3E238 A0A0K8UM71 A0A151JUJ4 A0A1L8HIJ1 A0A151K2H3 A0A0T6ATP1 T1I8D3 A0A1Y1NEC1 A0A151J2R5 W5UCF1 F4X4E7 Q6DCM0 A0A1S3ILL5 Q4S5M9 A0A3L8DKN7 A0A3B4DKX4 H2MBW0 F6VY14 H3D4W9 W5L5F6 A0A3P9L3Z7 A0A0P7XRK7 A0A3B3Z0K2 M4AQV5 A0A1W4XBH8 A0A158NL04 A0A3P9J828 A0A3B3BI55 Q6NW98 H2TBX3 A0A1A8HQS0 V9KNR2 A0A3P9NI22 A0A3B3RXD8 A0A3B3TNT5 A0A1A8L320 A0A1A8PLK6 G3NFR2 E2BTN0 A0A1A8BAE6 A0A087XX44 A0A1S3MD80 A0A1A8G9M0 A0A1A8PV98 A0A146WSR0 A0A060WRK9 A0A3P9DJQ9 A0A2D0QWR5 A0A1A8BJ87 A0A3B4VFI7 A0A2U9C0Y2 A0A3B4XGE5 I3KK17 A0A3P8YF51 A0A310S692 A0A1A7Z674 A0A3P8TN92 A0A2I4AJ78 A0A3B4GUU9 A0A026WM56 A0A0K8TLG4 A0A151X441 A0A3B5AX99 G3NFN3
A0A0N0P9W5 A0A194R5U0 A0A2H1VBA6 A0A067R739 E0VE24 A0A1B6C7V6 A0A1B6FTZ0 A0A1B6IPA1 A0A1B6LSC7 D6WNN3 A0A1Y1ND83 A0A0C9RF90 A0A1W4XLM8 A0A1S4F6T6 Q17DT7 A0A154PK45 A0A182GNX3 A0A182GCA7 A0A0L7R2H4 A0A087ZNG8 A0A1Q3EWE5 A0A0V0GAL8 B0XBT7 A0A069DU43 K7INI6 W8CBC7 A0A023EZB0 A0A1S3HFA3 A0A232FBH5 A0A034UZI2 A0A2A3E238 A0A0K8UM71 A0A151JUJ4 A0A1L8HIJ1 A0A151K2H3 A0A0T6ATP1 T1I8D3 A0A1Y1NEC1 A0A151J2R5 W5UCF1 F4X4E7 Q6DCM0 A0A1S3ILL5 Q4S5M9 A0A3L8DKN7 A0A3B4DKX4 H2MBW0 F6VY14 H3D4W9 W5L5F6 A0A3P9L3Z7 A0A0P7XRK7 A0A3B3Z0K2 M4AQV5 A0A1W4XBH8 A0A158NL04 A0A3P9J828 A0A3B3BI55 Q6NW98 H2TBX3 A0A1A8HQS0 V9KNR2 A0A3P9NI22 A0A3B3RXD8 A0A3B3TNT5 A0A1A8L320 A0A1A8PLK6 G3NFR2 E2BTN0 A0A1A8BAE6 A0A087XX44 A0A1S3MD80 A0A1A8G9M0 A0A1A8PV98 A0A146WSR0 A0A060WRK9 A0A3P9DJQ9 A0A2D0QWR5 A0A1A8BJ87 A0A3B4VFI7 A0A2U9C0Y2 A0A3B4XGE5 I3KK17 A0A3P8YF51 A0A310S692 A0A1A7Z674 A0A3P8TN92 A0A2I4AJ78 A0A3B4GUU9 A0A026WM56 A0A0K8TLG4 A0A151X441 A0A3B5AX99 G3NFN3
Pubmed
19121390
28756777
22118469
26227816
23622113
26354079
+ More
24845553 20566863 18362917 19820115 28004739 17510324 26483478 26334808 20075255 24495485 25474469 28648823 25348373 27762356 23127152 21719571 15496914 30249741 17554307 20431018 25329095 23542700 21347285 29451363 23594743 21551351 24402279 29240929 20798317 24755649 25186727 25069045 24508170 26369729
24845553 20566863 18362917 19820115 28004739 17510324 26483478 26334808 20075255 24495485 25474469 28648823 25348373 27762356 23127152 21719571 15496914 30249741 17554307 20431018 25329095 23542700 21347285 29451363 23594743 21551351 24402279 29240929 20798317 24755649 25186727 25069045 24508170 26369729
EMBL
BABH01027772
NWSH01004704
PCG64743.1
KZ150074
PZC73994.1
AGBW02009349
+ More
OWR51037.1 JTDY01008530 KOB64529.1 GAIX01005533 JAA87027.1 KQ459072 KPJ03838.1 KQ460685 KPJ12879.1 ODYU01001394 SOQ37514.1 KK852658 KDR19153.1 DS235088 EEB11630.1 GEDC01027978 GEDC01012229 JAS09320.1 JAS25069.1 GECZ01016125 JAS53644.1 GECU01018955 JAS88751.1 GEBQ01026458 GEBQ01013404 JAT13519.1 JAT26573.1 KQ971342 EFA03757.2 GEZM01009545 JAV94535.1 GBYB01002412 GBYB01015244 JAG72179.1 JAG85011.1 CH477291 EAT44575.1 KQ434938 KZC12167.1 JXUM01077310 KQ563000 KXJ74668.1 JXUM01054001 KQ561800 KXJ77495.1 KQ414666 KOC65077.1 GFDL01015420 JAV19625.1 GECL01001139 JAP04985.1 DS232653 EDS44466.1 GBGD01001256 JAC87633.1 GAMC01004936 JAC01620.1 GBBI01004616 JAC14096.1 NNAY01000558 OXU27677.1 GAKP01022531 JAC36421.1 KZ288438 PBC25755.1 GDHF01024525 JAI27789.1 KQ981746 KYN36174.1 CM004468 OCT95904.1 LKEX01012942 KYN50213.1 LJIG01022901 KRT78158.1 ACPB03001101 GEZM01009544 JAV94536.1 KQ980342 KYN16385.1 JT411880 AHH39643.1 GL888643 EGI58676.1 BC077988 AAH77988.1 CAAE01014729 CAG04053.1 QOIP01000007 RLU21015.1 AAMC01060383 JARO02000596 KPP78002.1 ADTU01019083 CABZ01067556 CABZ01067557 CU469570 BC067671 AAH67671.1 HAED01000592 HAEE01016076 SBQ86437.1 JW867404 AFO99921.1 HAEF01001404 SBR38786.1 HAEI01002803 HAEH01022190 SBR81894.1 GL450467 EFN80945.1 HADY01024975 HAEJ01011932 SBP63460.1 AYCK01008587 HAEB01021209 SBQ67736.1 HAEG01009703 SBR85181.1 GCES01054328 JAR31995.1 FR904684 CDQ69771.1 HADZ01003736 SBP67677.1 CP026253 AWP10275.1 AERX01002666 KQ768431 OAD53164.1 HADX01015614 SBP37846.1 KK107152 EZA57132.1 GDAI01002590 JAI15013.1 KQ982557 KYQ55039.1
OWR51037.1 JTDY01008530 KOB64529.1 GAIX01005533 JAA87027.1 KQ459072 KPJ03838.1 KQ460685 KPJ12879.1 ODYU01001394 SOQ37514.1 KK852658 KDR19153.1 DS235088 EEB11630.1 GEDC01027978 GEDC01012229 JAS09320.1 JAS25069.1 GECZ01016125 JAS53644.1 GECU01018955 JAS88751.1 GEBQ01026458 GEBQ01013404 JAT13519.1 JAT26573.1 KQ971342 EFA03757.2 GEZM01009545 JAV94535.1 GBYB01002412 GBYB01015244 JAG72179.1 JAG85011.1 CH477291 EAT44575.1 KQ434938 KZC12167.1 JXUM01077310 KQ563000 KXJ74668.1 JXUM01054001 KQ561800 KXJ77495.1 KQ414666 KOC65077.1 GFDL01015420 JAV19625.1 GECL01001139 JAP04985.1 DS232653 EDS44466.1 GBGD01001256 JAC87633.1 GAMC01004936 JAC01620.1 GBBI01004616 JAC14096.1 NNAY01000558 OXU27677.1 GAKP01022531 JAC36421.1 KZ288438 PBC25755.1 GDHF01024525 JAI27789.1 KQ981746 KYN36174.1 CM004468 OCT95904.1 LKEX01012942 KYN50213.1 LJIG01022901 KRT78158.1 ACPB03001101 GEZM01009544 JAV94536.1 KQ980342 KYN16385.1 JT411880 AHH39643.1 GL888643 EGI58676.1 BC077988 AAH77988.1 CAAE01014729 CAG04053.1 QOIP01000007 RLU21015.1 AAMC01060383 JARO02000596 KPP78002.1 ADTU01019083 CABZ01067556 CABZ01067557 CU469570 BC067671 AAH67671.1 HAED01000592 HAEE01016076 SBQ86437.1 JW867404 AFO99921.1 HAEF01001404 SBR38786.1 HAEI01002803 HAEH01022190 SBR81894.1 GL450467 EFN80945.1 HADY01024975 HAEJ01011932 SBP63460.1 AYCK01008587 HAEB01021209 SBQ67736.1 HAEG01009703 SBR85181.1 GCES01054328 JAR31995.1 FR904684 CDQ69771.1 HADZ01003736 SBP67677.1 CP026253 AWP10275.1 AERX01002666 KQ768431 OAD53164.1 HADX01015614 SBP37846.1 KK107152 EZA57132.1 GDAI01002590 JAI15013.1 KQ982557 KYQ55039.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053268
UP000053240
+ More
UP000027135 UP000009046 UP000007266 UP000192223 UP000008820 UP000076502 UP000069940 UP000249989 UP000053825 UP000005203 UP000002320 UP000002358 UP000085678 UP000215335 UP000242457 UP000078541 UP000186698 UP000078542 UP000015103 UP000078492 UP000221080 UP000007755 UP000279307 UP000261440 UP000001038 UP000008143 UP000007303 UP000018467 UP000265180 UP000034805 UP000192224 UP000261480 UP000002852 UP000005205 UP000265200 UP000261560 UP000000437 UP000005226 UP000242638 UP000261540 UP000261500 UP000007635 UP000008237 UP000028760 UP000087266 UP000265000 UP000193380 UP000265160 UP000261420 UP000246464 UP000261360 UP000005207 UP000265140 UP000265080 UP000192220 UP000261460 UP000053097 UP000075809 UP000261400
UP000027135 UP000009046 UP000007266 UP000192223 UP000008820 UP000076502 UP000069940 UP000249989 UP000053825 UP000005203 UP000002320 UP000002358 UP000085678 UP000215335 UP000242457 UP000078541 UP000186698 UP000078542 UP000015103 UP000078492 UP000221080 UP000007755 UP000279307 UP000261440 UP000001038 UP000008143 UP000007303 UP000018467 UP000265180 UP000034805 UP000192224 UP000261480 UP000002852 UP000005205 UP000265200 UP000261560 UP000000437 UP000005226 UP000242638 UP000261540 UP000261500 UP000007635 UP000008237 UP000028760 UP000087266 UP000265000 UP000193380 UP000265160 UP000261420 UP000246464 UP000261360 UP000005207 UP000265140 UP000265080 UP000192220 UP000261460 UP000053097 UP000075809 UP000261400
PRIDE
Pfam
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR024977 Apc4_WD40_dom
IPR019775 WD40_repeat_CS
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR036961 Kinesin_motor_dom_sf
IPR036640 ABC1_TM_sf
IPR019821 Kinesin_motor_CS
IPR001752 Kinesin_motor_dom
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR011527 ABC1_TM_dom
IPR003593 AAA+_ATPase
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR024977 Apc4_WD40_dom
IPR019775 WD40_repeat_CS
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR036961 Kinesin_motor_dom_sf
IPR036640 ABC1_TM_sf
IPR019821 Kinesin_motor_CS
IPR001752 Kinesin_motor_dom
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR011527 ABC1_TM_dom
IPR003593 AAA+_ATPase
Gene 3D
ProteinModelPortal
H9JD34
A0A2A4J051
A0A2W1BK21
A0A212FBE0
A0A0L7KNH6
S4PX74
+ More
A0A0N0P9W5 A0A194R5U0 A0A2H1VBA6 A0A067R739 E0VE24 A0A1B6C7V6 A0A1B6FTZ0 A0A1B6IPA1 A0A1B6LSC7 D6WNN3 A0A1Y1ND83 A0A0C9RF90 A0A1W4XLM8 A0A1S4F6T6 Q17DT7 A0A154PK45 A0A182GNX3 A0A182GCA7 A0A0L7R2H4 A0A087ZNG8 A0A1Q3EWE5 A0A0V0GAL8 B0XBT7 A0A069DU43 K7INI6 W8CBC7 A0A023EZB0 A0A1S3HFA3 A0A232FBH5 A0A034UZI2 A0A2A3E238 A0A0K8UM71 A0A151JUJ4 A0A1L8HIJ1 A0A151K2H3 A0A0T6ATP1 T1I8D3 A0A1Y1NEC1 A0A151J2R5 W5UCF1 F4X4E7 Q6DCM0 A0A1S3ILL5 Q4S5M9 A0A3L8DKN7 A0A3B4DKX4 H2MBW0 F6VY14 H3D4W9 W5L5F6 A0A3P9L3Z7 A0A0P7XRK7 A0A3B3Z0K2 M4AQV5 A0A1W4XBH8 A0A158NL04 A0A3P9J828 A0A3B3BI55 Q6NW98 H2TBX3 A0A1A8HQS0 V9KNR2 A0A3P9NI22 A0A3B3RXD8 A0A3B3TNT5 A0A1A8L320 A0A1A8PLK6 G3NFR2 E2BTN0 A0A1A8BAE6 A0A087XX44 A0A1S3MD80 A0A1A8G9M0 A0A1A8PV98 A0A146WSR0 A0A060WRK9 A0A3P9DJQ9 A0A2D0QWR5 A0A1A8BJ87 A0A3B4VFI7 A0A2U9C0Y2 A0A3B4XGE5 I3KK17 A0A3P8YF51 A0A310S692 A0A1A7Z674 A0A3P8TN92 A0A2I4AJ78 A0A3B4GUU9 A0A026WM56 A0A0K8TLG4 A0A151X441 A0A3B5AX99 G3NFN3
A0A0N0P9W5 A0A194R5U0 A0A2H1VBA6 A0A067R739 E0VE24 A0A1B6C7V6 A0A1B6FTZ0 A0A1B6IPA1 A0A1B6LSC7 D6WNN3 A0A1Y1ND83 A0A0C9RF90 A0A1W4XLM8 A0A1S4F6T6 Q17DT7 A0A154PK45 A0A182GNX3 A0A182GCA7 A0A0L7R2H4 A0A087ZNG8 A0A1Q3EWE5 A0A0V0GAL8 B0XBT7 A0A069DU43 K7INI6 W8CBC7 A0A023EZB0 A0A1S3HFA3 A0A232FBH5 A0A034UZI2 A0A2A3E238 A0A0K8UM71 A0A151JUJ4 A0A1L8HIJ1 A0A151K2H3 A0A0T6ATP1 T1I8D3 A0A1Y1NEC1 A0A151J2R5 W5UCF1 F4X4E7 Q6DCM0 A0A1S3ILL5 Q4S5M9 A0A3L8DKN7 A0A3B4DKX4 H2MBW0 F6VY14 H3D4W9 W5L5F6 A0A3P9L3Z7 A0A0P7XRK7 A0A3B3Z0K2 M4AQV5 A0A1W4XBH8 A0A158NL04 A0A3P9J828 A0A3B3BI55 Q6NW98 H2TBX3 A0A1A8HQS0 V9KNR2 A0A3P9NI22 A0A3B3RXD8 A0A3B3TNT5 A0A1A8L320 A0A1A8PLK6 G3NFR2 E2BTN0 A0A1A8BAE6 A0A087XX44 A0A1S3MD80 A0A1A8G9M0 A0A1A8PV98 A0A146WSR0 A0A060WRK9 A0A3P9DJQ9 A0A2D0QWR5 A0A1A8BJ87 A0A3B4VFI7 A0A2U9C0Y2 A0A3B4XGE5 I3KK17 A0A3P8YF51 A0A310S692 A0A1A7Z674 A0A3P8TN92 A0A2I4AJ78 A0A3B4GUU9 A0A026WM56 A0A0K8TLG4 A0A151X441 A0A3B5AX99 G3NFN3
PDB
4WJS
E-value=0.000290218,
Score=105
Ontologies
GO
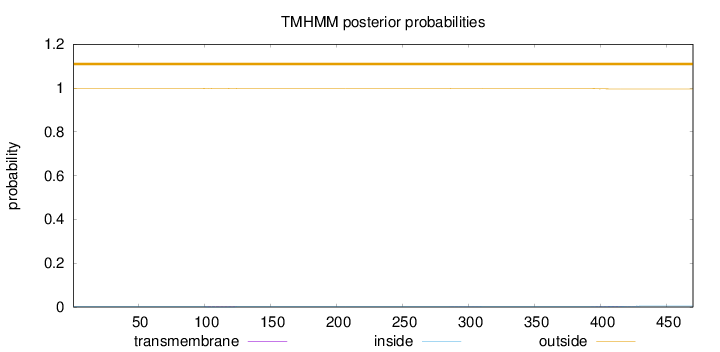
Topology
Length:
470
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10906
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00122
outside
1 - 470
Population Genetic Test Statistics
Pi
209.236876
Theta
177.518997
Tajima's D
0.918498
CLR
0.200554
CSRT
0.635818209089546
Interpretation
Uncertain
