Gene
KWMTBOMO01104
Pre Gene Modal
BGIBMGA007430
Annotation
putative_ubiquitin_specific_protease_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 4.074
Sequence
CDS
ATGAAAACTATTGGAAATGTAATAAATAAATACGATGTAACATTCGTTTTCAATGAAAATAAAGAAGATAATTTTAAATTTATGACTTTTATACCTCAAATTGATGAAGAAACTTGCAAAGAGATCGTAACGCTACCGAAAATAGTTAATATCTGTGCTAATGGCCAAATAGAAAGAAAAAGTATATTTCCATCGAAAAATCCGAAAGATATAAAAAAATGTCCTATTAATGTTGGCATGGGGTCTTTGTACCCTTTTGGAATTATTAATCACAAGGAGAAGTACAAAACGTTTGATCCACTGAACGAAACAGAAGTAAGAGGGTTGGATGTAGATTTAGTCAAAGTTCTAGTAAACCAGTTTAATGGTACATTGAATTTGTATTTCATCTATAAGAAAGAAGAAAATCCTTTTGGGCAATTGGATTTCATTCCTCTTGTACTTAATGGAAGCCTGGACGTCATTGCTGGCGGCTTTTACAGAATTTATGGAAATGTTGTGGCTTATTCCGGAATTTATACCAGTCAAGCCGTTACTTGGATGTATGTTGCAAACAGAACCACAAAATCCTGGCAAAGTCTAATAGTAAAAATAGACGGCTTATACATTTTTGTTATATTCCACCTCATATATAGCTATGTTTGGTACTTTGTGCGAAAATTTGATGAACAGGCAGTGGATTTTAGAAATACCATACTGTACAGTTGGGGAGCTTTGGTCGGCACTACATCGCTACAAGACGCTCTCTCTTTGAAACAGAGAATACTGAATTTAACGTACCTTATTATGTGTGTTCATTTATCAGCATACGTAAGTCTGCATTTATATTACTTCTTGACGGTTTTAGAGCCACCGGAGCTGTTGAAAAGTAATGATGACGTCATGCGATCCGGAAGACCCGCGTTCTTAATACCGATATCCAAATATTTCGTACTCGACGAGAAATACCTAAGTTTTGCTAACGCGTCCGAGGAATGCACGAAATTTCAGGACTGCTCCGACCTATCTCTACTTCGCAACGGCGTAACGATAATACTACAAGGATTCTTTTTGAATTACCAAGCGAGGACAGCTATTAACTATGAAGCTAAAGTACTCAGCGCTGCAGAGAACGTGTTAACTGTTTATTACGAGATGCTGTTGAGGAAGAACAGTCCCTACGTTGAAAGACTTCAGAAGCTGATGACACATTTGTTTGAAGCCGGCATACCAGATAGGTTTTATCGTCACGCTATTGGGTTGACGGTGATTGGGAAAGCTCACAGCGCCTGTCAGAACACAGTTAGCAACAGCTACAGTTGCCAGTCGGGCTGCAAAATAACTTTCGATCAATTCGCCGGAGTGTTTTATCTTTGGCTATTCGGATGTGTCTTGTCTTGTGGCGCTTTTATTTTCGAATTGTTCAGTAAATTCGGAAGGGCATAA
Protein
MKTIGNVINKYDVTFVFNENKEDNFKFMTFIPQIDEETCKEIVTLPKIVNICANGQIERKSIFPSKNPKDIKKCPINVGMGSLYPFGIINHKEKYKTFDPLNETEVRGLDVDLVKVLVNQFNGTLNLYFIYKKEENPFGQLDFIPLVLNGSLDVIAGGFYRIYGNVVAYSGIYTSQAVTWMYVANRTTKSWQSLIVKIDGLYIFVIFHLIYSYVWYFVRKFDEQAVDFRNTILYSWGALVGTTSLQDALSLKQRILNLTYLIMCVHLSAYVSLHLYYFLTVLEPPELLKSNDDVMRSGRPAFLIPISKYFVLDEKYLSFANASEECTKFQDCSDLSLLRNGVTIILQGFFLNYQARTAINYEAKVLSAAENVLTVYYEMLLRKNSPYVERLQKLMTHLFEAGIPDRFYRHAIGLTVIGKAHSACQNTVSNSYSCQSGCKITFDQFAGVFYLWLFGCVLSCGAFIFELFSKFGRA
Summary
Uniprot
H9JD33
A0A0L7KX39
A0A2W1BGQ0
A0A0N1PEI9
A0A2H1VPS6
A0A194R4Z7
+ More
A0A141W3J3 A0A141W3I4 A0A141W3G4 A0A141W3I8 A0A141W3J8 A0A141W3G9 A0A141W3H5 A0A140G9I3 A0A141W3K3 A0A212ELB3 A0A141W3H0 A0A141W3H9 A0A2P8XYF0 A0A067QXY6 A0A2P8YRK8 A0A2J7PQN4 A0A2J7PQL1 A0A2P8YDT9 A0A2P8ZGM7 D7EIG1 A0A067QL33 A0A067QJR5 A0A2J7Q9U6 T1HNN5 A0A2P8ZEC9 A0A2P8XGP6 J9LZH9 A0A2P8XYD1 A0A2P8Z120 A0A2P8YI71 A0A139W8Y9 A0A067R6W7 A0A336KKK9 A0A2P8XYR0 W5JW20 A0A2S2QMS9 A0A2P8YCF0 A0A2J7RBJ9 A0A2P8XKZ7 A0A2J7PQN8 A0A067RBW1 A0A2P8YDV8 A0A2P8XZE9 A0A2J7QXU2 A0A067QI43 A0A067RJB6 A0A2P8YI73 A0A067QYK3 A0A2J7PFF8 A0A2P8XI69 A0A2J7R371 A0A2J7PFZ6 A0A182PMW1 A0A067RWQ0 A0A067RC22 A0A2J7QT11 A0A084WIG5 A0A2P8XUX9 A0A067R490 A0A182H1I8 A0A1Q1PP69 A0A2J7PYQ2 A0A2P8XRP5 A0A2J7QJK4 A0A2P8ZEU7 A0A2J7RBK6 A0A2P8XZG1 A0A067R4T7 A0A2P8YN99 A0A2P8ZEH8 A0A2J7PZG5 A0A2J7QH68 A0A2J7PWK7 A0A2P8Y3B4 A0A067R8H5 A0A2J7QDG4 A0A3F2ZDG2 A0A2J7QC88 A0A2P8YJ37 A0A2P8YJH1 A0A2P8XTT7 A0A2P8ZA87 A0A2P8Y0F2 A0A2P8ZIG0 A0A2P8YDU3 A0A2P8XPB4 A0A2P8YR81 A0A2P8XPD0 A0A2J7QPS9
A0A141W3J3 A0A141W3I4 A0A141W3G4 A0A141W3I8 A0A141W3J8 A0A141W3G9 A0A141W3H5 A0A140G9I3 A0A141W3K3 A0A212ELB3 A0A141W3H0 A0A141W3H9 A0A2P8XYF0 A0A067QXY6 A0A2P8YRK8 A0A2J7PQN4 A0A2J7PQL1 A0A2P8YDT9 A0A2P8ZGM7 D7EIG1 A0A067QL33 A0A067QJR5 A0A2J7Q9U6 T1HNN5 A0A2P8ZEC9 A0A2P8XGP6 J9LZH9 A0A2P8XYD1 A0A2P8Z120 A0A2P8YI71 A0A139W8Y9 A0A067R6W7 A0A336KKK9 A0A2P8XYR0 W5JW20 A0A2S2QMS9 A0A2P8YCF0 A0A2J7RBJ9 A0A2P8XKZ7 A0A2J7PQN8 A0A067RBW1 A0A2P8YDV8 A0A2P8XZE9 A0A2J7QXU2 A0A067QI43 A0A067RJB6 A0A2P8YI73 A0A067QYK3 A0A2J7PFF8 A0A2P8XI69 A0A2J7R371 A0A2J7PFZ6 A0A182PMW1 A0A067RWQ0 A0A067RC22 A0A2J7QT11 A0A084WIG5 A0A2P8XUX9 A0A067R490 A0A182H1I8 A0A1Q1PP69 A0A2J7PYQ2 A0A2P8XRP5 A0A2J7QJK4 A0A2P8ZEU7 A0A2J7RBK6 A0A2P8XZG1 A0A067R4T7 A0A2P8YN99 A0A2P8ZEH8 A0A2J7PZG5 A0A2J7QH68 A0A2J7PWK7 A0A2P8Y3B4 A0A067R8H5 A0A2J7QDG4 A0A3F2ZDG2 A0A2J7QC88 A0A2P8YJ37 A0A2P8YJH1 A0A2P8XTT7 A0A2P8ZA87 A0A2P8Y0F2 A0A2P8ZIG0 A0A2P8YDU3 A0A2P8XPB4 A0A2P8YR81 A0A2P8XPD0 A0A2J7QPS9
Pubmed
EMBL
BABH01027764
JTDY01004899
KOB67619.1
KZ150074
PZC73988.1
KQ459072
+ More
KPJ03839.1 ODYU01003550 SOQ42462.1 KQ460685 KPJ12878.1 KU756967 AMM70700.1 KU756958 AMM70691.1 KU756938 AMM70671.1 KU756962 AMM70695.1 KU756972 AMM70705.1 KU756943 AMM70676.1 KU756949 AMM70682.1 KU702635 AMM70662.1 KU756977 AMM70710.1 AGBW02014103 OWR42257.1 KU756944 AMM70677.1 KU756953 AMM70686.1 PYGN01001167 PSN37025.1 KK853123 KDR11041.1 PYGN01000408 PSN46889.1 NEVH01022635 PNF18649.1 PNF18624.1 PYGN01000674 PSN42422.1 PYGN01000063 PSN55655.1 DS497678 EFA11918.1 KK853197 KDR09956.1 KK853275 KDR09085.1 NEVH01016344 PNF25358.1 ACPB03001833 PYGN01000081 PSN54862.1 PYGN01002203 PSN31178.1 ABLF02041229 PSN37021.1 PYGN01000247 PSN50195.1 PYGN01000578 PSN43951.1 KQ972879 KXZ75746.1 KK852902 KDR14037.1 UFQS01000221 UFQT01000221 SSX01560.1 SSX21940.1 PYGN01001151 PSN37148.1 ADMH02000120 ETN67703.1 GGMS01009876 MBY79079.1 PYGN01000706 PSN41928.1 NEVH01005895 PNF38217.1 PYGN01001824 PSN32652.1 PNF18650.1 KK852566 KDR21232.1 PSN42426.1 PYGN01001126 PSN37378.1 NEVH01009372 PNF33402.1 KK853359 KDR08169.1 KK852437 KDR23912.1 PSN43950.1 KK852871 KDR14577.1 NEVH01025660 PNF15065.1 PYGN01002051 PSN31682.1 NEVH01007822 PNF35288.1 NEVH01025635 PNF15248.1 KK852417 KDR24339.1 KK852744 KDR17342.1 NEVH01011202 PNF31732.1 ATLV01023931 KE525347 KFB50009.1 PYGN01001306 PSN35821.1 KDR14038.1 JXUM01023173 KQ560653 KXJ81430.1 KY325458 AQM73619.1 NEVH01020342 PNF21465.1 PYGN01001465 PSN34682.1 NEVH01013553 PNF28758.1 PYGN01000077 PSN55016.1 PNF38215.1 PSN37383.1 KK852749 KDR17222.1 PYGN01000473 PSN45721.1 PSN54860.1 NEVH01020335 PNF21729.1 NEVH01014356 PNF27931.1 NEVH01020889 PNF20710.1 PYGN01000978 PSN38746.1 PSN38753.1 KK852669 KDR18856.1 NEVH01015822 PNF26625.1 AJWK01002246 NEVH01016291 PNF26182.1 PYGN01000561 PSN44214.1 PYGN01000552 PSN44386.1 PYGN01001360 PSN35409.1 PYGN01000127 PSN53423.1 PYGN01001086 PSN37735.1 PYGN01000048 PSN56284.1 PSN42427.1 PYGN01001589 PSN33849.1 PYGN01000417 PSN46760.1 PSN33850.1 NEVH01012087 PNF30598.1
KPJ03839.1 ODYU01003550 SOQ42462.1 KQ460685 KPJ12878.1 KU756967 AMM70700.1 KU756958 AMM70691.1 KU756938 AMM70671.1 KU756962 AMM70695.1 KU756972 AMM70705.1 KU756943 AMM70676.1 KU756949 AMM70682.1 KU702635 AMM70662.1 KU756977 AMM70710.1 AGBW02014103 OWR42257.1 KU756944 AMM70677.1 KU756953 AMM70686.1 PYGN01001167 PSN37025.1 KK853123 KDR11041.1 PYGN01000408 PSN46889.1 NEVH01022635 PNF18649.1 PNF18624.1 PYGN01000674 PSN42422.1 PYGN01000063 PSN55655.1 DS497678 EFA11918.1 KK853197 KDR09956.1 KK853275 KDR09085.1 NEVH01016344 PNF25358.1 ACPB03001833 PYGN01000081 PSN54862.1 PYGN01002203 PSN31178.1 ABLF02041229 PSN37021.1 PYGN01000247 PSN50195.1 PYGN01000578 PSN43951.1 KQ972879 KXZ75746.1 KK852902 KDR14037.1 UFQS01000221 UFQT01000221 SSX01560.1 SSX21940.1 PYGN01001151 PSN37148.1 ADMH02000120 ETN67703.1 GGMS01009876 MBY79079.1 PYGN01000706 PSN41928.1 NEVH01005895 PNF38217.1 PYGN01001824 PSN32652.1 PNF18650.1 KK852566 KDR21232.1 PSN42426.1 PYGN01001126 PSN37378.1 NEVH01009372 PNF33402.1 KK853359 KDR08169.1 KK852437 KDR23912.1 PSN43950.1 KK852871 KDR14577.1 NEVH01025660 PNF15065.1 PYGN01002051 PSN31682.1 NEVH01007822 PNF35288.1 NEVH01025635 PNF15248.1 KK852417 KDR24339.1 KK852744 KDR17342.1 NEVH01011202 PNF31732.1 ATLV01023931 KE525347 KFB50009.1 PYGN01001306 PSN35821.1 KDR14038.1 JXUM01023173 KQ560653 KXJ81430.1 KY325458 AQM73619.1 NEVH01020342 PNF21465.1 PYGN01001465 PSN34682.1 NEVH01013553 PNF28758.1 PYGN01000077 PSN55016.1 PNF38215.1 PSN37383.1 KK852749 KDR17222.1 PYGN01000473 PSN45721.1 PSN54860.1 NEVH01020335 PNF21729.1 NEVH01014356 PNF27931.1 NEVH01020889 PNF20710.1 PYGN01000978 PSN38746.1 PSN38753.1 KK852669 KDR18856.1 NEVH01015822 PNF26625.1 AJWK01002246 NEVH01016291 PNF26182.1 PYGN01000561 PSN44214.1 PYGN01000552 PSN44386.1 PYGN01001360 PSN35409.1 PYGN01000127 PSN53423.1 PYGN01001086 PSN37735.1 PYGN01000048 PSN56284.1 PSN42427.1 PYGN01001589 PSN33849.1 PYGN01000417 PSN46760.1 PSN33850.1 NEVH01012087 PNF30598.1
Proteomes
PRIDE
Interpro
CDD
ProteinModelPortal
H9JD33
A0A0L7KX39
A0A2W1BGQ0
A0A0N1PEI9
A0A2H1VPS6
A0A194R4Z7
+ More
A0A141W3J3 A0A141W3I4 A0A141W3G4 A0A141W3I8 A0A141W3J8 A0A141W3G9 A0A141W3H5 A0A140G9I3 A0A141W3K3 A0A212ELB3 A0A141W3H0 A0A141W3H9 A0A2P8XYF0 A0A067QXY6 A0A2P8YRK8 A0A2J7PQN4 A0A2J7PQL1 A0A2P8YDT9 A0A2P8ZGM7 D7EIG1 A0A067QL33 A0A067QJR5 A0A2J7Q9U6 T1HNN5 A0A2P8ZEC9 A0A2P8XGP6 J9LZH9 A0A2P8XYD1 A0A2P8Z120 A0A2P8YI71 A0A139W8Y9 A0A067R6W7 A0A336KKK9 A0A2P8XYR0 W5JW20 A0A2S2QMS9 A0A2P8YCF0 A0A2J7RBJ9 A0A2P8XKZ7 A0A2J7PQN8 A0A067RBW1 A0A2P8YDV8 A0A2P8XZE9 A0A2J7QXU2 A0A067QI43 A0A067RJB6 A0A2P8YI73 A0A067QYK3 A0A2J7PFF8 A0A2P8XI69 A0A2J7R371 A0A2J7PFZ6 A0A182PMW1 A0A067RWQ0 A0A067RC22 A0A2J7QT11 A0A084WIG5 A0A2P8XUX9 A0A067R490 A0A182H1I8 A0A1Q1PP69 A0A2J7PYQ2 A0A2P8XRP5 A0A2J7QJK4 A0A2P8ZEU7 A0A2J7RBK6 A0A2P8XZG1 A0A067R4T7 A0A2P8YN99 A0A2P8ZEH8 A0A2J7PZG5 A0A2J7QH68 A0A2J7PWK7 A0A2P8Y3B4 A0A067R8H5 A0A2J7QDG4 A0A3F2ZDG2 A0A2J7QC88 A0A2P8YJ37 A0A2P8YJH1 A0A2P8XTT7 A0A2P8ZA87 A0A2P8Y0F2 A0A2P8ZIG0 A0A2P8YDU3 A0A2P8XPB4 A0A2P8YR81 A0A2P8XPD0 A0A2J7QPS9
A0A141W3J3 A0A141W3I4 A0A141W3G4 A0A141W3I8 A0A141W3J8 A0A141W3G9 A0A141W3H5 A0A140G9I3 A0A141W3K3 A0A212ELB3 A0A141W3H0 A0A141W3H9 A0A2P8XYF0 A0A067QXY6 A0A2P8YRK8 A0A2J7PQN4 A0A2J7PQL1 A0A2P8YDT9 A0A2P8ZGM7 D7EIG1 A0A067QL33 A0A067QJR5 A0A2J7Q9U6 T1HNN5 A0A2P8ZEC9 A0A2P8XGP6 J9LZH9 A0A2P8XYD1 A0A2P8Z120 A0A2P8YI71 A0A139W8Y9 A0A067R6W7 A0A336KKK9 A0A2P8XYR0 W5JW20 A0A2S2QMS9 A0A2P8YCF0 A0A2J7RBJ9 A0A2P8XKZ7 A0A2J7PQN8 A0A067RBW1 A0A2P8YDV8 A0A2P8XZE9 A0A2J7QXU2 A0A067QI43 A0A067RJB6 A0A2P8YI73 A0A067QYK3 A0A2J7PFF8 A0A2P8XI69 A0A2J7R371 A0A2J7PFZ6 A0A182PMW1 A0A067RWQ0 A0A067RC22 A0A2J7QT11 A0A084WIG5 A0A2P8XUX9 A0A067R490 A0A182H1I8 A0A1Q1PP69 A0A2J7PYQ2 A0A2P8XRP5 A0A2J7QJK4 A0A2P8ZEU7 A0A2J7RBK6 A0A2P8XZG1 A0A067R4T7 A0A2P8YN99 A0A2P8ZEH8 A0A2J7PZG5 A0A2J7QH68 A0A2J7PWK7 A0A2P8Y3B4 A0A067R8H5 A0A2J7QDG4 A0A3F2ZDG2 A0A2J7QC88 A0A2P8YJ37 A0A2P8YJH1 A0A2P8XTT7 A0A2P8ZA87 A0A2P8Y0F2 A0A2P8ZIG0 A0A2P8YDU3 A0A2P8XPB4 A0A2P8YR81 A0A2P8XPD0 A0A2J7QPS9
Ontologies
KEGG
GO
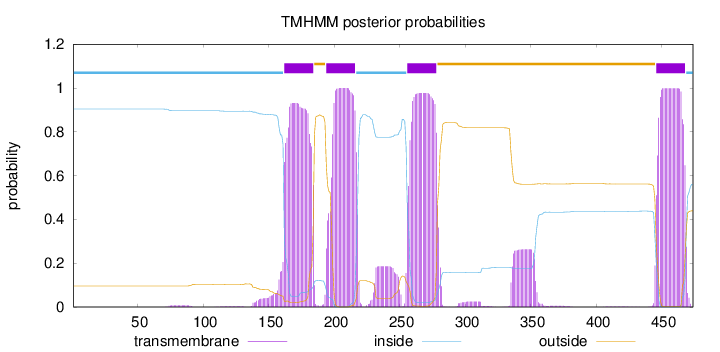
Topology
Length:
474
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
95.5017500000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.90380
inside
1 - 161
TMhelix
162 - 184
outside
185 - 193
TMhelix
194 - 216
inside
217 - 255
TMhelix
256 - 278
outside
279 - 445
TMhelix
446 - 468
inside
469 - 474
Population Genetic Test Statistics
Pi
249.115855
Theta
221.264705
Tajima's D
0.385236
CLR
0.129029
CSRT
0.47957602119894
Interpretation
Uncertain
