Gene
KWMTBOMO01100
Annotation
PREDICTED:_rho-related_GTP-binding_protein_RhoN-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.158 Extracellular Reliability : 1.679
Sequence
CDS
ATGTGGTCGAATAGAAAATGCGAAACCGGTACAGCTGGTACTCCTACAGCAGAAGAAGAGATCAAAGTAGTCCTTGTAGGAGATACTCAGTGTGGGAAGACGTCGCTGGTCCAAAGATTTGTGTCCGATACATTTATTGAGGCCTACACTCCGACCGGCTTCGAGAAGTACAGCTACACATGCTCAGTGGCCGATTACAAAGTAAACTTTTCAATCTGGGATACGTCCGGAACGACGTCCTACGACACGGTCAGACCTTTAGCGTACCAAGACGCGAAGCTGTTTATGCTGTGCTTCAACGTAGCCGAACCTGAGACCTTACACAACGCCGCAGCCAAATGGTACAAGGAGGTGAGGACCCACGGTGGCTCAGCGCCTGTGGTTCTGTGCGGATGCAAAGCGGACCTGAGACACGACAAGGAAACGCTCACAGTTCTGTCCAAACTAAAAACACATCCAGTCACCAGTGAAGAGTAG
Protein
MWSNRKCETGTAGTPTAEEEIKVVLVGDTQCGKTSLVQRFVSDTFIEAYTPTGFEKYSYTCSVADYKVNFSIWDTSGTTSYDTVRPLAYQDAKLFMLCFNVAEPETLHNAAAKWYKEVRTHGGSAPVVLCGCKADLRHDKETLTVLSKLKTHPVTSEE
Summary
Uniprot
A0A212FJ52
A0A194R6K8
A0A2H1V979
A0A2A4JEL7
A0A2P8Z5H8
A0A0N1IN71
+ More
A0A1B6CR53 A0A2J7PI92 A0A067QJ14 A0A1W7R9T0 T1JA45 T1JA46 A0A087SZL0 K1QCI4 V5GVZ3 B7P1J6 A0A131XVL3 A0A293M309 A0A1Z5L7L5 A0A2R5LKR0 A0A1E1XKQ6 A0A0K8RNM7 A0A1E1X8G6 A0A224Z010 L7MFP7 A0A0N9HHJ4 A0A0N9HGZ5 C3YW53 V3ZMI5 A0A2D0S7T4 A0A2T7P8L3 A0A3B4DT00 A0A3P8XQQ9 A0A3B1JG08 A0A3B4DZ49 H3AP85 A0A3P9KJZ8 A0A3P9H1M9 W5UE43 A0A3B3H890 A0A2D4FCE1 A0A3P8WI60 A0A1S3NFK8 A0A3B5AXM9 I3IX41 A0A3B4G3S8 A0A3P8NBX6 A0A3P9D8K5 A0A0B7A8L5 A0A3N0Y237 A0A2I4CDW9 A0A2C9LHN7 G1KNT4 F1RCR8 Q0P4B0 A0A3B3VGS3 A0A3P9NS29 A0A3B5Q347 A0A1W5AWK4 W5N058 A0A087XT01 G3P321 A0A151PC09 A0A1U7RVF4 A0A3P8YNC6 A0A3P9AA65 H3BB75 A0A1A7WKW7 A0A1A8RK84 A0A1A8QIT7 A0A1A8E1Y4 A0A1A8FQ43 A0A1A8V9F3 A0A2U9CM92 A0A3B4VPK7 H0YXC3 A0A3B3ZEF9 A0A3P8RXS3 A0A1S3R848 H2LIC6 A0A3P9IAG4 A0A0B7A6C1 A0A2U9CT72 A0A0P6JDM6 A0A3B3D1B3 A0A146W456 A0A3B3BJ30 A0A1S3GF04 A0A286XMT5 A0A3B3DYV4 H3AP86 H2UY56 H2MA67 I3LQJ8 A0A3P8VFY0 A0A0S7L170 A0A2P4TG55 A0A226MJ23 A0A3M0JKQ7 A0A226NUX2
A0A1B6CR53 A0A2J7PI92 A0A067QJ14 A0A1W7R9T0 T1JA45 T1JA46 A0A087SZL0 K1QCI4 V5GVZ3 B7P1J6 A0A131XVL3 A0A293M309 A0A1Z5L7L5 A0A2R5LKR0 A0A1E1XKQ6 A0A0K8RNM7 A0A1E1X8G6 A0A224Z010 L7MFP7 A0A0N9HHJ4 A0A0N9HGZ5 C3YW53 V3ZMI5 A0A2D0S7T4 A0A2T7P8L3 A0A3B4DT00 A0A3P8XQQ9 A0A3B1JG08 A0A3B4DZ49 H3AP85 A0A3P9KJZ8 A0A3P9H1M9 W5UE43 A0A3B3H890 A0A2D4FCE1 A0A3P8WI60 A0A1S3NFK8 A0A3B5AXM9 I3IX41 A0A3B4G3S8 A0A3P8NBX6 A0A3P9D8K5 A0A0B7A8L5 A0A3N0Y237 A0A2I4CDW9 A0A2C9LHN7 G1KNT4 F1RCR8 Q0P4B0 A0A3B3VGS3 A0A3P9NS29 A0A3B5Q347 A0A1W5AWK4 W5N058 A0A087XT01 G3P321 A0A151PC09 A0A1U7RVF4 A0A3P8YNC6 A0A3P9AA65 H3BB75 A0A1A7WKW7 A0A1A8RK84 A0A1A8QIT7 A0A1A8E1Y4 A0A1A8FQ43 A0A1A8V9F3 A0A2U9CM92 A0A3B4VPK7 H0YXC3 A0A3B3ZEF9 A0A3P8RXS3 A0A1S3R848 H2LIC6 A0A3P9IAG4 A0A0B7A6C1 A0A2U9CT72 A0A0P6JDM6 A0A3B3D1B3 A0A146W456 A0A3B3BJ30 A0A1S3GF04 A0A286XMT5 A0A3B3DYV4 H3AP86 H2UY56 H2MA67 I3LQJ8 A0A3P8VFY0 A0A0S7L170 A0A2P4TG55 A0A226MJ23 A0A3M0JKQ7 A0A226NUX2
Pubmed
EMBL
AGBW02008315
OWR53761.1
KQ460685
KPJ12875.1
ODYU01001335
SOQ37377.1
+ More
NWSH01001863 PCG69880.1 PYGN01000186 PSN51753.1 KQ459072 KPJ03843.1 GEDC01021411 JAS15887.1 NEVH01025128 PNF16060.1 KK853461 KDR07480.1 GFAH01000489 JAV47900.1 JH431984 KK112695 KFM58299.1 JH817257 EKC19221.1 GANP01009923 JAB74545.1 ABJB010491955 ABJB010744779 ABJB010797205 ABJB010901470 ABJB010961825 DS616900 EEC00468.1 GEFM01005548 JAP70248.1 GFWV01008646 MAA33375.1 GFJQ02003611 JAW03359.1 GGLE01005977 MBY10103.1 GFAA01003650 JAT99784.1 GADI01001157 JAA72651.1 GFAC01003633 JAT95555.1 GFPF01008204 MAA19350.1 GACK01001998 JAA63036.1 KT037725 KT037743 ALG00090.1 KT037734 KT037752 NEDP02003109 ALG00117.1 OWF49441.1 GG666559 EEN55507.1 KB204047 ESO82046.1 PZQS01000005 PVD29746.1 AFYH01167400 AFYH01167401 AFYH01167402 AFYH01167403 AFYH01167404 JT407398 AHH37972.1 IACJ01060618 LAA44896.1 AERX01007585 HACG01029465 CEK76330.1 RJVU01053643 ROL27933.1 BX572620 CR855865 BX901909 BX936331 BC122188 BC165068 AAI22189.1 AAI65068.1 AHAT01022968 AHAT01022969 AYCK01007245 AKHW03000499 KYO46469.1 AFYH01034171 AFYH01034172 AFYH01034173 AFYH01034174 HADW01004841 HADX01010466 SBP06241.1 HAEH01015199 HAEI01008717 SBS05783.1 HAEF01007803 HAEG01012696 SBR93014.1 HADZ01011103 HAEA01011503 SBQ39983.1 HAEB01014369 HAEC01005629 SBQ60896.1 HADY01017202 HAEJ01015750 SBS56207.1 CP026260 AWP17744.1 ABQF01052718 ABQF01052719 ABQF01052720 ABQF01052721 HACG01029463 CEK76328.1 CP026261 AWP19323.1 GEBF01001544 JAO02089.1 GCES01062514 JAR23809.1 AAKN02042423 AEMK02000080 GBYX01189704 JAO83280.1 PPHD01000557 POI35336.1 MCFN01000771 OXB55282.1 QRBI01000137 RMC01513.1 AWGT02000662 OXB71112.1
NWSH01001863 PCG69880.1 PYGN01000186 PSN51753.1 KQ459072 KPJ03843.1 GEDC01021411 JAS15887.1 NEVH01025128 PNF16060.1 KK853461 KDR07480.1 GFAH01000489 JAV47900.1 JH431984 KK112695 KFM58299.1 JH817257 EKC19221.1 GANP01009923 JAB74545.1 ABJB010491955 ABJB010744779 ABJB010797205 ABJB010901470 ABJB010961825 DS616900 EEC00468.1 GEFM01005548 JAP70248.1 GFWV01008646 MAA33375.1 GFJQ02003611 JAW03359.1 GGLE01005977 MBY10103.1 GFAA01003650 JAT99784.1 GADI01001157 JAA72651.1 GFAC01003633 JAT95555.1 GFPF01008204 MAA19350.1 GACK01001998 JAA63036.1 KT037725 KT037743 ALG00090.1 KT037734 KT037752 NEDP02003109 ALG00117.1 OWF49441.1 GG666559 EEN55507.1 KB204047 ESO82046.1 PZQS01000005 PVD29746.1 AFYH01167400 AFYH01167401 AFYH01167402 AFYH01167403 AFYH01167404 JT407398 AHH37972.1 IACJ01060618 LAA44896.1 AERX01007585 HACG01029465 CEK76330.1 RJVU01053643 ROL27933.1 BX572620 CR855865 BX901909 BX936331 BC122188 BC165068 AAI22189.1 AAI65068.1 AHAT01022968 AHAT01022969 AYCK01007245 AKHW03000499 KYO46469.1 AFYH01034171 AFYH01034172 AFYH01034173 AFYH01034174 HADW01004841 HADX01010466 SBP06241.1 HAEH01015199 HAEI01008717 SBS05783.1 HAEF01007803 HAEG01012696 SBR93014.1 HADZ01011103 HAEA01011503 SBQ39983.1 HAEB01014369 HAEC01005629 SBQ60896.1 HADY01017202 HAEJ01015750 SBS56207.1 CP026260 AWP17744.1 ABQF01052718 ABQF01052719 ABQF01052720 ABQF01052721 HACG01029463 CEK76328.1 CP026261 AWP19323.1 GEBF01001544 JAO02089.1 GCES01062514 JAR23809.1 AAKN02042423 AEMK02000080 GBYX01189704 JAO83280.1 PPHD01000557 POI35336.1 MCFN01000771 OXB55282.1 QRBI01000137 RMC01513.1 AWGT02000662 OXB71112.1
Proteomes
UP000007151
UP000053240
UP000218220
UP000245037
UP000053268
UP000235965
+ More
UP000027135 UP000054359 UP000005408 UP000001555 UP000242188 UP000001554 UP000030746 UP000221080 UP000245119 UP000261440 UP000265140 UP000018467 UP000008672 UP000265180 UP000265200 UP000001038 UP000265120 UP000087266 UP000261400 UP000005207 UP000261460 UP000265100 UP000265160 UP000192220 UP000076420 UP000001646 UP000000437 UP000261500 UP000242638 UP000002852 UP000192224 UP000018468 UP000028760 UP000007635 UP000050525 UP000189705 UP000246464 UP000261420 UP000007754 UP000261520 UP000265080 UP000261560 UP000081671 UP000005447 UP000005226 UP000008227 UP000198323 UP000269221 UP000198419
UP000027135 UP000054359 UP000005408 UP000001555 UP000242188 UP000001554 UP000030746 UP000221080 UP000245119 UP000261440 UP000265140 UP000018467 UP000008672 UP000265180 UP000265200 UP000001038 UP000265120 UP000087266 UP000261400 UP000005207 UP000261460 UP000265100 UP000265160 UP000192220 UP000076420 UP000001646 UP000000437 UP000261500 UP000242638 UP000002852 UP000192224 UP000018468 UP000028760 UP000007635 UP000050525 UP000189705 UP000246464 UP000261420 UP000007754 UP000261520 UP000265080 UP000261560 UP000081671 UP000005447 UP000005226 UP000008227 UP000198323 UP000269221 UP000198419
PRIDE
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A212FJ52
A0A194R6K8
A0A2H1V979
A0A2A4JEL7
A0A2P8Z5H8
A0A0N1IN71
+ More
A0A1B6CR53 A0A2J7PI92 A0A067QJ14 A0A1W7R9T0 T1JA45 T1JA46 A0A087SZL0 K1QCI4 V5GVZ3 B7P1J6 A0A131XVL3 A0A293M309 A0A1Z5L7L5 A0A2R5LKR0 A0A1E1XKQ6 A0A0K8RNM7 A0A1E1X8G6 A0A224Z010 L7MFP7 A0A0N9HHJ4 A0A0N9HGZ5 C3YW53 V3ZMI5 A0A2D0S7T4 A0A2T7P8L3 A0A3B4DT00 A0A3P8XQQ9 A0A3B1JG08 A0A3B4DZ49 H3AP85 A0A3P9KJZ8 A0A3P9H1M9 W5UE43 A0A3B3H890 A0A2D4FCE1 A0A3P8WI60 A0A1S3NFK8 A0A3B5AXM9 I3IX41 A0A3B4G3S8 A0A3P8NBX6 A0A3P9D8K5 A0A0B7A8L5 A0A3N0Y237 A0A2I4CDW9 A0A2C9LHN7 G1KNT4 F1RCR8 Q0P4B0 A0A3B3VGS3 A0A3P9NS29 A0A3B5Q347 A0A1W5AWK4 W5N058 A0A087XT01 G3P321 A0A151PC09 A0A1U7RVF4 A0A3P8YNC6 A0A3P9AA65 H3BB75 A0A1A7WKW7 A0A1A8RK84 A0A1A8QIT7 A0A1A8E1Y4 A0A1A8FQ43 A0A1A8V9F3 A0A2U9CM92 A0A3B4VPK7 H0YXC3 A0A3B3ZEF9 A0A3P8RXS3 A0A1S3R848 H2LIC6 A0A3P9IAG4 A0A0B7A6C1 A0A2U9CT72 A0A0P6JDM6 A0A3B3D1B3 A0A146W456 A0A3B3BJ30 A0A1S3GF04 A0A286XMT5 A0A3B3DYV4 H3AP86 H2UY56 H2MA67 I3LQJ8 A0A3P8VFY0 A0A0S7L170 A0A2P4TG55 A0A226MJ23 A0A3M0JKQ7 A0A226NUX2
A0A1B6CR53 A0A2J7PI92 A0A067QJ14 A0A1W7R9T0 T1JA45 T1JA46 A0A087SZL0 K1QCI4 V5GVZ3 B7P1J6 A0A131XVL3 A0A293M309 A0A1Z5L7L5 A0A2R5LKR0 A0A1E1XKQ6 A0A0K8RNM7 A0A1E1X8G6 A0A224Z010 L7MFP7 A0A0N9HHJ4 A0A0N9HGZ5 C3YW53 V3ZMI5 A0A2D0S7T4 A0A2T7P8L3 A0A3B4DT00 A0A3P8XQQ9 A0A3B1JG08 A0A3B4DZ49 H3AP85 A0A3P9KJZ8 A0A3P9H1M9 W5UE43 A0A3B3H890 A0A2D4FCE1 A0A3P8WI60 A0A1S3NFK8 A0A3B5AXM9 I3IX41 A0A3B4G3S8 A0A3P8NBX6 A0A3P9D8K5 A0A0B7A8L5 A0A3N0Y237 A0A2I4CDW9 A0A2C9LHN7 G1KNT4 F1RCR8 Q0P4B0 A0A3B3VGS3 A0A3P9NS29 A0A3B5Q347 A0A1W5AWK4 W5N058 A0A087XT01 G3P321 A0A151PC09 A0A1U7RVF4 A0A3P8YNC6 A0A3P9AA65 H3BB75 A0A1A7WKW7 A0A1A8RK84 A0A1A8QIT7 A0A1A8E1Y4 A0A1A8FQ43 A0A1A8V9F3 A0A2U9CM92 A0A3B4VPK7 H0YXC3 A0A3B3ZEF9 A0A3P8RXS3 A0A1S3R848 H2LIC6 A0A3P9IAG4 A0A0B7A6C1 A0A2U9CT72 A0A0P6JDM6 A0A3B3D1B3 A0A146W456 A0A3B3BJ30 A0A1S3GF04 A0A286XMT5 A0A3B3DYV4 H3AP86 H2UY56 H2MA67 I3LQJ8 A0A3P8VFY0 A0A0S7L170 A0A2P4TG55 A0A226MJ23 A0A3M0JKQ7 A0A226NUX2
PDB
2J1L
E-value=6.67556e-31,
Score=329
Ontologies
GO
Topology
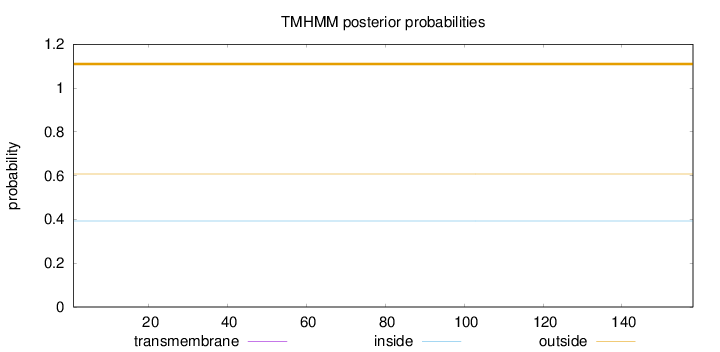
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00102
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.39268
outside
1 - 158
Population Genetic Test Statistics
Pi
181.141761
Theta
147.838547
Tajima's D
0.782633
CLR
0.458372
CSRT
0.593770311484426
Interpretation
Uncertain
