Gene
KWMTBOMO01097
Annotation
PREDICTED:_lysozyme_2-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.084
Sequence
CDS
ATGTTTGTTACGAGATCGTTAGTATTTCCTGCTTGTTTCGTTGCAGTCTTCGTGTTTTGGATAACCGGTTCAGGAGGAGTTTTCATTTCAAATTTGTCGGAGAGCTGCTTCAGATGCCTCTGCTACGTATCCACAAAATGTAATCTTGCACACGACTGTACTGGAGGATACTGTGGACCATTTAATATATCCAAGGTGTATTGGGTGGATGCAGGAAAGGTTGTGCACTCCGAAGATACCCCGGAGAGGAGCCATGCCTGGGAAGACTGCGCAAGAAACTATCATTGCTCAAAGAAAATAATCGAAGGCTATTTAGAACGGTTTGGAAAGGACTGCAACGGTGATGGCGTGACGGACTGCTTCGACTACATGATGGTGAACGGTAACGGTGGGTACGGTTGCACCAAGCCCCTTAACCGCTCAGTCAACGGACGACGGTGGCTTCTGCGCTACGAAGAGTGCACTTTATAA
Protein
MFVTRSLVFPACFVAVFVFWITGSGGVFISNLSESCFRCLCYVSTKCNLAHDCTGGYCGPFNISKVYWVDAGKVVHSEDTPERSHAWEDCARNYHCSKKIIEGYLERFGKDCNGDGVTDCFDYMMVNGNGGYGCTKPLNRSVNGRRWLLRYEECTL
Summary
Similarity
Belongs to the insulin family.
Uniprot
A0A2H1WZL5
A0A194R5B8
A0A2W1BB65
A0A346RAE5
A0A0N0PA83
A0A212F5L8
+ More
A0A212ESN9 I4DRR6 A0A194QA92 A0A2H1X1S6 A0A0N1IPP7 A0A3G1KL21 B6RQP9 A0A139WE07 A0A0S1TPE9 Q17L20 A0A182H5V7 A0A182HBT7 A0A1Q3FR03 A0A2M3ZC39 A0A067RKT7 A0A182PSU9 A0A1B6CQ25 B0WQ70 A0A2M4AY59 A0A2M4AY31 A0A182KBM7 A0A084VNW1 A0A1B6LYK0 A0A1W4W6S3 A0A2M4DQI9 A0A182MNV1 A0A182N8G3 A0A182VF42 A0A182QAL9 A0A182W5M4 A0A182XBY8 A0A182TX19 A0A182RHL0 A0A182IAJ8 A0A182L877 A0A1S3DJS5 Q7QF28 A0A3Q0JJK3 A0A182SGQ0 A0A1B6IEC7 A0A2M4C2I4 A0A2S2QVX7 J9K6K4 A0A1Z1MYP5 A0A2H8TR34 M9ZV91 A0A212F6D5 A0A1Y1KC06 A0A2S2P8F2 A0A1B6FY81 A0A1W5RN49 E9GN48 A0A164RN61 A0A0P5HTN5 A0A0P5LQP0 A0A0P6BZH4 A0A0P6GVU8 A0A0S1TQ24 A0A0P5J1D4 A0A023EG44 A0A023EJ95 E0W1V9 A0A0T6AYW8 B4LP77 A0A1D2N4H7 B4LP76 A0A139WE63
A0A212ESN9 I4DRR6 A0A194QA92 A0A2H1X1S6 A0A0N1IPP7 A0A3G1KL21 B6RQP9 A0A139WE07 A0A0S1TPE9 Q17L20 A0A182H5V7 A0A182HBT7 A0A1Q3FR03 A0A2M3ZC39 A0A067RKT7 A0A182PSU9 A0A1B6CQ25 B0WQ70 A0A2M4AY59 A0A2M4AY31 A0A182KBM7 A0A084VNW1 A0A1B6LYK0 A0A1W4W6S3 A0A2M4DQI9 A0A182MNV1 A0A182N8G3 A0A182VF42 A0A182QAL9 A0A182W5M4 A0A182XBY8 A0A182TX19 A0A182RHL0 A0A182IAJ8 A0A182L877 A0A1S3DJS5 Q7QF28 A0A3Q0JJK3 A0A182SGQ0 A0A1B6IEC7 A0A2M4C2I4 A0A2S2QVX7 J9K6K4 A0A1Z1MYP5 A0A2H8TR34 M9ZV91 A0A212F6D5 A0A1Y1KC06 A0A2S2P8F2 A0A1B6FY81 A0A1W5RN49 E9GN48 A0A164RN61 A0A0P5HTN5 A0A0P5LQP0 A0A0P6BZH4 A0A0P6GVU8 A0A0S1TQ24 A0A0P5J1D4 A0A023EG44 A0A023EJ95 E0W1V9 A0A0T6AYW8 B4LP77 A0A1D2N4H7 B4LP76 A0A139WE63
Pubmed
EMBL
ODYU01012294
SOQ58529.1
KQ460685
KPJ12872.1
KZ150177
PZC72562.1
+ More
MH200935 AXS59145.1 KQ459072 KPJ03846.1 AGBW02010175 OWR49030.1 AGBW02012762 OWR44497.1 AK405241 BAM20606.1 KQ459249 KPJ02349.1 ODYU01012789 SOQ59273.1 KQ460366 KPJ15588.1 KY986535 ATU47289.1 EU282120 ABZ80670.1 KQ971357 KYB26180.1 KT380886 ALM25916.1 CH477219 EAT47407.1 JXUM01026700 KQ560764 KXJ80981.1 JXUM01032636 KQ560962 KXJ80191.1 GFDL01005107 JAV29938.1 GGFM01005321 MBW26072.1 KK852618 KDR20128.1 GEDC01021682 GEDC01015235 JAS15616.1 JAS22063.1 DS232036 EDS32730.1 GGFK01012389 MBW45710.1 GGFK01012388 MBW45709.1 ATLV01014931 KE524996 KFB39655.1 GEBQ01011218 GEBQ01003431 JAT28759.1 JAT36546.1 GGFL01015639 MBW79817.1 AXCM01002712 AXCN02002226 APCN01001259 EF492429 AAAB01008846 ABP35929.1 EAA06475.5 GECU01022463 JAS85243.1 GGFJ01010381 MBW59522.1 GGMS01012714 MBY81917.1 ABLF02035118 ABLF02035120 KY575341 ARW71317.1 GFXV01004654 MBW16459.1 KC355205 AGK40905.1 AGBW02010050 OWR49284.1 GEZM01090935 JAV57155.1 GGMR01013110 MBY25729.1 GECZ01014619 JAS55150.1 KY113350 AQV03739.1 GL732554 EFX79098.1 LRGB01002190 KZS08800.1 GDIQ01228318 JAK23407.1 GDIQ01166592 JAK85133.1 GDIP01007032 JAM96683.1 GDIQ01028774 JAN65963.1 KT380887 ALM25917.1 GDIQ01231020 JAK20705.1 JXUM01064974 JXUM01064975 JXUM01064976 JXUM01064977 JXUM01064978 JXUM01064979 JXUM01086087 GAPW01005281 KQ563543 KQ562324 JAC08317.1 KXJ73684.1 KXJ76142.1 GAPW01004567 JAC09031.1 DS235873 EEB19553.1 LJIG01022494 KRT80280.1 CH940648 EDW60186.2 LJIJ01000228 ODN00174.1 EDW60185.1 KYB26179.1
MH200935 AXS59145.1 KQ459072 KPJ03846.1 AGBW02010175 OWR49030.1 AGBW02012762 OWR44497.1 AK405241 BAM20606.1 KQ459249 KPJ02349.1 ODYU01012789 SOQ59273.1 KQ460366 KPJ15588.1 KY986535 ATU47289.1 EU282120 ABZ80670.1 KQ971357 KYB26180.1 KT380886 ALM25916.1 CH477219 EAT47407.1 JXUM01026700 KQ560764 KXJ80981.1 JXUM01032636 KQ560962 KXJ80191.1 GFDL01005107 JAV29938.1 GGFM01005321 MBW26072.1 KK852618 KDR20128.1 GEDC01021682 GEDC01015235 JAS15616.1 JAS22063.1 DS232036 EDS32730.1 GGFK01012389 MBW45710.1 GGFK01012388 MBW45709.1 ATLV01014931 KE524996 KFB39655.1 GEBQ01011218 GEBQ01003431 JAT28759.1 JAT36546.1 GGFL01015639 MBW79817.1 AXCM01002712 AXCN02002226 APCN01001259 EF492429 AAAB01008846 ABP35929.1 EAA06475.5 GECU01022463 JAS85243.1 GGFJ01010381 MBW59522.1 GGMS01012714 MBY81917.1 ABLF02035118 ABLF02035120 KY575341 ARW71317.1 GFXV01004654 MBW16459.1 KC355205 AGK40905.1 AGBW02010050 OWR49284.1 GEZM01090935 JAV57155.1 GGMR01013110 MBY25729.1 GECZ01014619 JAS55150.1 KY113350 AQV03739.1 GL732554 EFX79098.1 LRGB01002190 KZS08800.1 GDIQ01228318 JAK23407.1 GDIQ01166592 JAK85133.1 GDIP01007032 JAM96683.1 GDIQ01028774 JAN65963.1 KT380887 ALM25917.1 GDIQ01231020 JAK20705.1 JXUM01064974 JXUM01064975 JXUM01064976 JXUM01064977 JXUM01064978 JXUM01064979 JXUM01086087 GAPW01005281 KQ563543 KQ562324 JAC08317.1 KXJ73684.1 KXJ76142.1 GAPW01004567 JAC09031.1 DS235873 EEB19553.1 LJIG01022494 KRT80280.1 CH940648 EDW60186.2 LJIJ01000228 ODN00174.1 EDW60185.1 KYB26179.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000007266
UP000008820
UP000069940
+ More
UP000249989 UP000027135 UP000075885 UP000002320 UP000075881 UP000030765 UP000192223 UP000075883 UP000075884 UP000075903 UP000075886 UP000075920 UP000076407 UP000075902 UP000075900 UP000075840 UP000075882 UP000079169 UP000007062 UP000075901 UP000007819 UP000000305 UP000076858 UP000009046 UP000008792 UP000094527
UP000249989 UP000027135 UP000075885 UP000002320 UP000075881 UP000030765 UP000192223 UP000075883 UP000075884 UP000075903 UP000075886 UP000075920 UP000076407 UP000075902 UP000075900 UP000075840 UP000075882 UP000079169 UP000007062 UP000075901 UP000007819 UP000000305 UP000076858 UP000009046 UP000008792 UP000094527
Interpro
ProteinModelPortal
A0A2H1WZL5
A0A194R5B8
A0A2W1BB65
A0A346RAE5
A0A0N0PA83
A0A212F5L8
+ More
A0A212ESN9 I4DRR6 A0A194QA92 A0A2H1X1S6 A0A0N1IPP7 A0A3G1KL21 B6RQP9 A0A139WE07 A0A0S1TPE9 Q17L20 A0A182H5V7 A0A182HBT7 A0A1Q3FR03 A0A2M3ZC39 A0A067RKT7 A0A182PSU9 A0A1B6CQ25 B0WQ70 A0A2M4AY59 A0A2M4AY31 A0A182KBM7 A0A084VNW1 A0A1B6LYK0 A0A1W4W6S3 A0A2M4DQI9 A0A182MNV1 A0A182N8G3 A0A182VF42 A0A182QAL9 A0A182W5M4 A0A182XBY8 A0A182TX19 A0A182RHL0 A0A182IAJ8 A0A182L877 A0A1S3DJS5 Q7QF28 A0A3Q0JJK3 A0A182SGQ0 A0A1B6IEC7 A0A2M4C2I4 A0A2S2QVX7 J9K6K4 A0A1Z1MYP5 A0A2H8TR34 M9ZV91 A0A212F6D5 A0A1Y1KC06 A0A2S2P8F2 A0A1B6FY81 A0A1W5RN49 E9GN48 A0A164RN61 A0A0P5HTN5 A0A0P5LQP0 A0A0P6BZH4 A0A0P6GVU8 A0A0S1TQ24 A0A0P5J1D4 A0A023EG44 A0A023EJ95 E0W1V9 A0A0T6AYW8 B4LP77 A0A1D2N4H7 B4LP76 A0A139WE63
A0A212ESN9 I4DRR6 A0A194QA92 A0A2H1X1S6 A0A0N1IPP7 A0A3G1KL21 B6RQP9 A0A139WE07 A0A0S1TPE9 Q17L20 A0A182H5V7 A0A182HBT7 A0A1Q3FR03 A0A2M3ZC39 A0A067RKT7 A0A182PSU9 A0A1B6CQ25 B0WQ70 A0A2M4AY59 A0A2M4AY31 A0A182KBM7 A0A084VNW1 A0A1B6LYK0 A0A1W4W6S3 A0A2M4DQI9 A0A182MNV1 A0A182N8G3 A0A182VF42 A0A182QAL9 A0A182W5M4 A0A182XBY8 A0A182TX19 A0A182RHL0 A0A182IAJ8 A0A182L877 A0A1S3DJS5 Q7QF28 A0A3Q0JJK3 A0A182SGQ0 A0A1B6IEC7 A0A2M4C2I4 A0A2S2QVX7 J9K6K4 A0A1Z1MYP5 A0A2H8TR34 M9ZV91 A0A212F6D5 A0A1Y1KC06 A0A2S2P8F2 A0A1B6FY81 A0A1W5RN49 E9GN48 A0A164RN61 A0A0P5HTN5 A0A0P5LQP0 A0A0P6BZH4 A0A0P6GVU8 A0A0S1TQ24 A0A0P5J1D4 A0A023EG44 A0A023EJ95 E0W1V9 A0A0T6AYW8 B4LP77 A0A1D2N4H7 B4LP76 A0A139WE63
PDB
4PJ2
E-value=0.0027723,
Score=90
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 25,
Likelihood: 0.887030
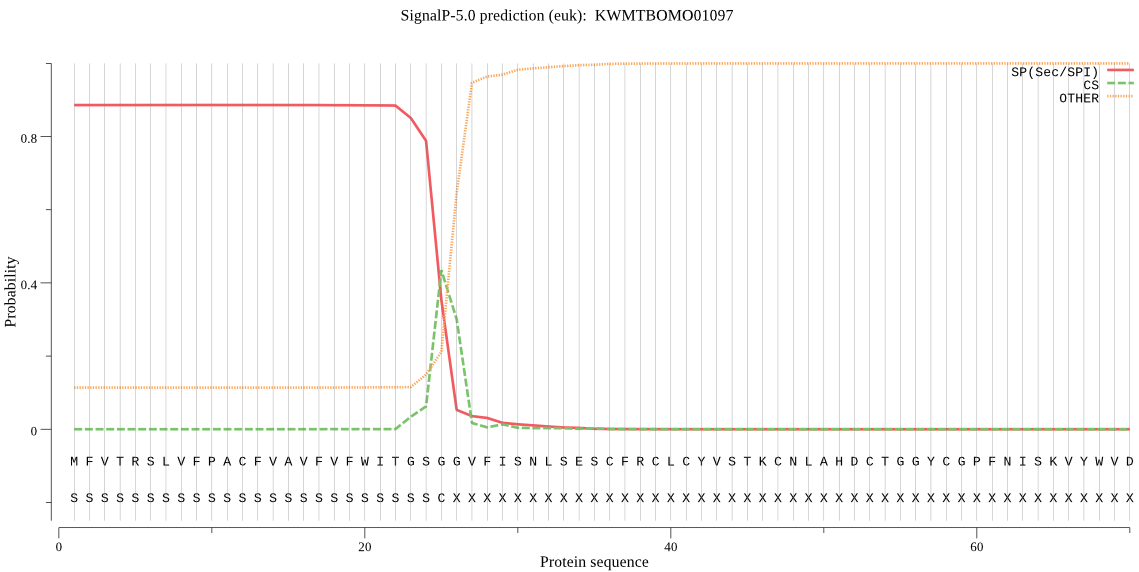
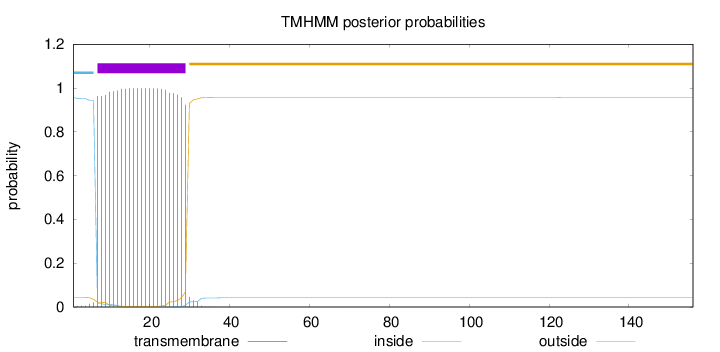
Length:
156
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.78974
Exp number, first 60 AAs:
22.78699
Total prob of N-in:
0.95662
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 156
Population Genetic Test Statistics
Pi
42.166289
Theta
166.06992
Tajima's D
0.025472
CLR
1.738545
CSRT
0.379381030948453
Interpretation
Uncertain
