Gene
KWMTBOMO01094
Pre Gene Modal
BGIBMGA007342
Annotation
PREDICTED:_protein_bicaudal_C_homolog_1-A_[Bombyx_mori]
Full name
Protein bicaudal C
Location in the cell
Mitochondrial Reliability : 1.217 Nuclear Reliability : 1.715
Sequence
CDS
ATGGCTACGACCAATTACTTAACGGTAAAAACGCATTTATCAGAATTTAAAACAGACAAAGATGCATTAAGTGAGGCGTCAGAAAGCGGTACAAGTAACTCGGTGACGTCAGCGGAAGACCTACAAAAATTAGCCGTAATACTCGGTTTGAATAGTGCCGAGGATGTCTACCAAGAGAGATTTCGAGTTGACAGACGACGCCTTGAGGCTATGTTGGGTGATAACCTCGAGGAATCACAAACAGCGCAAGCATTCTTTCATCGGGTAATGAATGAAACGAACACTTTAATAAATTGGCCTGCACGCTTGAAAATCGGAGCTCGCTCTAAAAAAGATCCTCACGTGAGAGTCGCTGGTCGACCTGATGATGTGAAATTGGCTCGTGAAAAAATTATGCAGTTACTGGACACAAGGAGTAATCGTGTTACAATGAAAATCGACGTCAGCTACACGGACCATTCGCACATAATCGGCAAGGGTGGTTTGACGATAAAACGCGTCATGGAAGAGACGGGCTGCCACATTCACTTCCCGGACTCGAACAGGACCAGCACCGTGGAGAAGAGTAACCAGGTCTCCATCGCAGGTGATATGGAAAGAGTCGAACGAGCCAGGGCAAGGGTTCGAGCGCTAACCCCACTAGTATTCAGCTTTGAGCTTCCGATAGTGTCGGCTTCACAACCGCTGCCGGACATTAACTCGCCCTACGTACATCAGATACAAGAACAGTATAACGTTCAGGTGATGATCCGTAACCGTCCGAAGCTCCACGCCAACCTGCTGGTGGTGAAGGGAGTCCAGTGGGAAGTGCAGGCCACCATGGAGGCCACCAAACTACTCATGAACTACATGTGCGGACCACTCGCTAGCCAAACCCTGGTCCAGATGTCACTGGAAATATCTCCGATGCACCACGCCGTGGTGGTAGGTCGGGGTGCTGAGCAGCTGAAGGTCATCATGAAGGGCAGCGGGACGCAGATAATGTTCCCGGACGCGGACGACCCCAACATTCCGCCCCTCAAAAAGAGCTGCGTCACTATCACCGGACAGATCCAAGACGTGTACACGGCCAGGCAACAACTTGTGGGCAGCCTGCCGCTGGTGGTGATCTTCGATGTGTCTGACGAGGCGTGTCGCGTGGACAACGACGCCGCCAACAAGCTGATGCAGAAACACAACGTCTACATCAACATCAGACGCAAGCCTAAGCAGGGGATCGCTTCCGTCGTCATCAAGGGGATAGAGAGATGCGCGGGCGACATCTATGAAGCCCGCCGCGAGCTACTGGGCGGGAAGGAGCCGGTCATCGCAGCGGAGATACCGGAGTCGTACAACATTCCGTCGCTGGGCAGCACCATGCCCAACTTCGGGCCGTTCAGTCCGTTCTCGGCGCACGCGTCGCCCTACAGCCCCGAGTGCGGCGGTGCGGCGGGGCCCGCGCCCTACACGCCCGAGTACGGCGCCGTGGTGTCGCCCGGCGCGTGGCTGCTCAGCCCCCCGGCGAGGGCGGCGCACTTCCACTACCCCGCGCTGGGACCGACCCTACAGCTGCCGTCACAGAACTCCGTCGTGTACAAGAATCAAACTTGGAACCGTACGTTTTTTTTATTATTGCTTAGATGA
Protein
MATTNYLTVKTHLSEFKTDKDALSEASESGTSNSVTSAEDLQKLAVILGLNSAEDVYQERFRVDRRRLEAMLGDNLEESQTAQAFFHRVMNETNTLINWPARLKIGARSKKDPHVRVAGRPDDVKLAREKIMQLLDTRSNRVTMKIDVSYTDHSHIIGKGGLTIKRVMEETGCHIHFPDSNRTSTVEKSNQVSIAGDMERVERARARVRALTPLVFSFELPIVSASQPLPDINSPYVHQIQEQYNVQVMIRNRPKLHANLLVVKGVQWEVQATMEATKLLMNYMCGPLASQTLVQMSLEISPMHHAVVVGRGAEQLKVIMKGSGTQIMFPDADDPNIPPLKKSCVTITGQIQDVYTARQQLVGSLPLVVIFDVSDEACRVDNDAANKLMQKHNVYINIRRKPKQGIASVVIKGIERCAGDIYEARRELLGGKEPVIAAEIPESYNIPSLGSTMPNFGPFSPFSAHASPYSPECGGAAGPAPYTPEYGAVVSPGAWLLSPPARAAHFHYPALGPTLQLPSQNSVVYKNQTWNRTFFLLLLR
Summary
Description
RNA-binding protein that is involved in oogenesis. Required for correct targeting of the migrating anterior follicle cells and the establishment of anterior-posterior polarity in the oocyte. May act as translational repressor of oskar during oogenesis. Function seems to be sensitive to small changes in expression.
Similarity
Belongs to the BicC family.
Keywords
Complete proteome
Developmental protein
Differentiation
Oogenesis
Phosphoprotein
Reference proteome
Repeat
RNA-binding
Feature
chain Protein bicaudal C
Uniprot
S4PKQ2
A0A212FKQ6
A0A2W1BAV0
A0A0N1I581
A0A194R4Y5
A0A0L7R4Y6
+ More
E2B2B9 A0A2A3EFG7 A0A195D177 A0A195B6E3 A0A158NH18 A0A151XDL7 A0A151J205 A0A195FNS9 A0A026VZM3 A0A067RDT9 A0A2A4J394 A0A1B6DKN9 A0A2J7QGK9 A0A310SKW2 A0A1Y1LKG0 K7IRE5 A0A2S2N6L7 A0A2P8XRN2 A0A2S2NW32 A0A2H8TX19 A0A2S2Q3V5 A0A1B6KZ36 A0A232F179 A0A023F1U7 D6WV91 E0VH60 B0WFE1 A0A1Q3EVJ0 U5EZ73 A0A182NU51 A0A084WG14 A0A182J0X5 A0A182L6E4 A0A182RIN9 A0A1S4H1E8 A0A182YDQ3 Q7PVI4 A0A182HGH0 A0A1I8M2W9 A0A182UVZ3 A0A182X0T4 A0A182QH92 Q16TR2 A0A182F3K6 A0A182VUH0 A0A182GRC3 A0A182MG49 A0A0P6K0N0 A0A182K9R6 A0A336LJF4 A0A034VR21 A0A0A1XF30 W5JCR6 A0A182P4D3 A0A0L0C5B3 A0A0P4Y0B1 A0A0K8VVZ4 A0A0K8VQH4 A0A0P5D1P4 A0A0P6GTX1 A0A0P5X730 A0A0P5ACE1 B3N639 E9HKF0 A0A336K4K9 A0A0P6H0P0 B4I1Z4 A0A0J9TP72 B4Q6X9 E2QCR4 Q24009 A0A0P5RDT7 B4NXR7 A0A3B0K8S0 A0A0P5RKI6 A0A0K8UTE3 A0A0K8V9P2 A0A1J1IL71 Q29KC7 A0A1W4WAQ7 B3MKY9 J9K7C8 A0A0M4ERR7 A0A0M4ESA7
E2B2B9 A0A2A3EFG7 A0A195D177 A0A195B6E3 A0A158NH18 A0A151XDL7 A0A151J205 A0A195FNS9 A0A026VZM3 A0A067RDT9 A0A2A4J394 A0A1B6DKN9 A0A2J7QGK9 A0A310SKW2 A0A1Y1LKG0 K7IRE5 A0A2S2N6L7 A0A2P8XRN2 A0A2S2NW32 A0A2H8TX19 A0A2S2Q3V5 A0A1B6KZ36 A0A232F179 A0A023F1U7 D6WV91 E0VH60 B0WFE1 A0A1Q3EVJ0 U5EZ73 A0A182NU51 A0A084WG14 A0A182J0X5 A0A182L6E4 A0A182RIN9 A0A1S4H1E8 A0A182YDQ3 Q7PVI4 A0A182HGH0 A0A1I8M2W9 A0A182UVZ3 A0A182X0T4 A0A182QH92 Q16TR2 A0A182F3K6 A0A182VUH0 A0A182GRC3 A0A182MG49 A0A0P6K0N0 A0A182K9R6 A0A336LJF4 A0A034VR21 A0A0A1XF30 W5JCR6 A0A182P4D3 A0A0L0C5B3 A0A0P4Y0B1 A0A0K8VVZ4 A0A0K8VQH4 A0A0P5D1P4 A0A0P6GTX1 A0A0P5X730 A0A0P5ACE1 B3N639 E9HKF0 A0A336K4K9 A0A0P6H0P0 B4I1Z4 A0A0J9TP72 B4Q6X9 E2QCR4 Q24009 A0A0P5RDT7 B4NXR7 A0A3B0K8S0 A0A0P5RKI6 A0A0K8UTE3 A0A0K8V9P2 A0A1J1IL71 Q29KC7 A0A1W4WAQ7 B3MKY9 J9K7C8 A0A0M4ERR7 A0A0M4ESA7
Pubmed
23622113
22118469
28756777
26354079
20798317
21347285
+ More
24508170 30249741 24845553 28004739 20075255 29403074 28648823 25474469 18362917 19820115 20566863 24438588 20966253 25244985 12364791 25315136 17510324 26483478 26999592 25348373 25830018 20920257 23761445 26108605 17994087 21292972 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7538070 12537569 9671494 18327897 17550304 15632085 18057021
24508170 30249741 24845553 28004739 20075255 29403074 28648823 25474469 18362917 19820115 20566863 24438588 20966253 25244985 12364791 25315136 17510324 26483478 26999592 25348373 25830018 20920257 23761445 26108605 17994087 21292972 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7538070 12537569 9671494 18327897 17550304 15632085 18057021
EMBL
GAIX01004380
JAA88180.1
AGBW02008002
OWR54324.1
KZ150177
PZC72559.1
+ More
KQ459072 KPJ03849.1 KQ460685 KPJ12868.1 KQ414654 KOC65945.1 GL445130 EFN90156.1 KZ288256 PBC30533.1 KQ976973 KYN06665.1 KQ976574 KYM80088.1 ADTU01015436 ADTU01015437 ADTU01015438 ADTU01015439 KQ982294 KYQ58348.1 KQ980447 KYN16015.1 KQ981387 KYN41967.1 KK107566 QOIP01000006 EZA48916.1 RLU22042.1 KK852699 KDR18180.1 NWSH01003336 PCG66525.1 GEDC01011042 JAS26256.1 NEVH01014361 PNF27721.1 KQ761783 OAD57009.1 GEZM01053183 JAV74139.1 GGMR01000186 MBY12805.1 PYGN01001467 PSN34659.1 GGMR01008814 MBY21433.1 GFXV01006992 MBW18797.1 GGMS01003203 MBY72406.1 GEBQ01023306 JAT16671.1 NNAY01001342 OXU24300.1 GBBI01003726 JAC14986.1 KQ971363 EFA07761.1 DS235161 EEB12716.1 DS231917 EDS26180.1 GFDL01015817 JAV19228.1 GANO01001311 JAB58560.1 ATLV01023428 KE525343 KFB49158.1 AAAB01008984 EAA14677.4 APCN01004228 AXCN02000773 CH477642 EAT37905.1 JXUM01082123 JXUM01082124 KQ563292 KXJ74114.1 AXCM01000083 GDUN01001111 JAN94808.1 UFQS01000023 UFQT01000023 SSW97634.1 SSX18020.1 GAKP01014030 GAKP01014029 GAKP01014028 GAKP01014027 JAC44924.1 GBXI01005777 GBXI01004293 JAD08515.1 JAD09999.1 ADMH02001769 ETN61133.1 JRES01000973 KNC26629.1 GDIP01234320 JAI89081.1 GDHF01009261 GDHF01008243 GDHF01000569 JAI43053.1 JAI44071.1 JAI51745.1 GDHF01019225 GDHF01012514 GDHF01011173 JAI33089.1 JAI39800.1 JAI41141.1 GDIP01162379 JAJ61023.1 GDIQ01029715 JAN65022.1 GDIP01076437 JAM27278.1 GDIP01201880 JAJ21522.1 CH954177 EDV58077.1 GL732668 EFX67775.1 SSW97633.1 SSX18019.1 GDIQ01026401 JAN68336.1 CH480820 EDW54551.1 CM002910 KMY90450.1 CM000361 EDX05187.1 AE014134 AAF53499.1 U15928 BT023837 GDIQ01103869 JAL47857.1 CM000157 EDW89692.1 KRJ98664.1 OUUW01000006 SPP82459.1 GDIQ01105778 JAL45948.1 GDHF01024674 GDHF01023373 GDHF01022352 GDHF01013749 GDHF01004800 GDHF01004285 GDHF01002889 JAI27640.1 JAI28941.1 JAI29962.1 JAI38565.1 JAI47514.1 JAI48029.1 JAI49425.1 GDHF01016771 GDHF01014204 JAI35543.1 JAI38110.1 CVRI01000055 CRL00989.1 CH379061 EAL33248.1 CH902620 EDV30647.1 KPU72974.1 KPU72975.1 ABLF02031103 CP012523 ALC39912.1 ALC39920.1
KQ459072 KPJ03849.1 KQ460685 KPJ12868.1 KQ414654 KOC65945.1 GL445130 EFN90156.1 KZ288256 PBC30533.1 KQ976973 KYN06665.1 KQ976574 KYM80088.1 ADTU01015436 ADTU01015437 ADTU01015438 ADTU01015439 KQ982294 KYQ58348.1 KQ980447 KYN16015.1 KQ981387 KYN41967.1 KK107566 QOIP01000006 EZA48916.1 RLU22042.1 KK852699 KDR18180.1 NWSH01003336 PCG66525.1 GEDC01011042 JAS26256.1 NEVH01014361 PNF27721.1 KQ761783 OAD57009.1 GEZM01053183 JAV74139.1 GGMR01000186 MBY12805.1 PYGN01001467 PSN34659.1 GGMR01008814 MBY21433.1 GFXV01006992 MBW18797.1 GGMS01003203 MBY72406.1 GEBQ01023306 JAT16671.1 NNAY01001342 OXU24300.1 GBBI01003726 JAC14986.1 KQ971363 EFA07761.1 DS235161 EEB12716.1 DS231917 EDS26180.1 GFDL01015817 JAV19228.1 GANO01001311 JAB58560.1 ATLV01023428 KE525343 KFB49158.1 AAAB01008984 EAA14677.4 APCN01004228 AXCN02000773 CH477642 EAT37905.1 JXUM01082123 JXUM01082124 KQ563292 KXJ74114.1 AXCM01000083 GDUN01001111 JAN94808.1 UFQS01000023 UFQT01000023 SSW97634.1 SSX18020.1 GAKP01014030 GAKP01014029 GAKP01014028 GAKP01014027 JAC44924.1 GBXI01005777 GBXI01004293 JAD08515.1 JAD09999.1 ADMH02001769 ETN61133.1 JRES01000973 KNC26629.1 GDIP01234320 JAI89081.1 GDHF01009261 GDHF01008243 GDHF01000569 JAI43053.1 JAI44071.1 JAI51745.1 GDHF01019225 GDHF01012514 GDHF01011173 JAI33089.1 JAI39800.1 JAI41141.1 GDIP01162379 JAJ61023.1 GDIQ01029715 JAN65022.1 GDIP01076437 JAM27278.1 GDIP01201880 JAJ21522.1 CH954177 EDV58077.1 GL732668 EFX67775.1 SSW97633.1 SSX18019.1 GDIQ01026401 JAN68336.1 CH480820 EDW54551.1 CM002910 KMY90450.1 CM000361 EDX05187.1 AE014134 AAF53499.1 U15928 BT023837 GDIQ01103869 JAL47857.1 CM000157 EDW89692.1 KRJ98664.1 OUUW01000006 SPP82459.1 GDIQ01105778 JAL45948.1 GDHF01024674 GDHF01023373 GDHF01022352 GDHF01013749 GDHF01004800 GDHF01004285 GDHF01002889 JAI27640.1 JAI28941.1 JAI29962.1 JAI38565.1 JAI47514.1 JAI48029.1 JAI49425.1 GDHF01016771 GDHF01014204 JAI35543.1 JAI38110.1 CVRI01000055 CRL00989.1 CH379061 EAL33248.1 CH902620 EDV30647.1 KPU72974.1 KPU72975.1 ABLF02031103 CP012523 ALC39912.1 ALC39920.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000053825
UP000008237
UP000242457
+ More
UP000078542 UP000078540 UP000005205 UP000075809 UP000078492 UP000078541 UP000053097 UP000279307 UP000027135 UP000218220 UP000235965 UP000002358 UP000245037 UP000215335 UP000007266 UP000009046 UP000002320 UP000075884 UP000030765 UP000075880 UP000075882 UP000075900 UP000076408 UP000007062 UP000075840 UP000095301 UP000075903 UP000076407 UP000075886 UP000008820 UP000069272 UP000075920 UP000069940 UP000249989 UP000075883 UP000075881 UP000000673 UP000075885 UP000037069 UP000008711 UP000000305 UP000001292 UP000000304 UP000000803 UP000002282 UP000268350 UP000183832 UP000001819 UP000192221 UP000007801 UP000007819 UP000092553
UP000078542 UP000078540 UP000005205 UP000075809 UP000078492 UP000078541 UP000053097 UP000279307 UP000027135 UP000218220 UP000235965 UP000002358 UP000245037 UP000215335 UP000007266 UP000009046 UP000002320 UP000075884 UP000030765 UP000075880 UP000075882 UP000075900 UP000076408 UP000007062 UP000075840 UP000095301 UP000075903 UP000076407 UP000075886 UP000008820 UP000069272 UP000075920 UP000069940 UP000249989 UP000075883 UP000075881 UP000000673 UP000075885 UP000037069 UP000008711 UP000000305 UP000001292 UP000000304 UP000000803 UP000002282 UP000268350 UP000183832 UP000001819 UP000192221 UP000007801 UP000007819 UP000092553
Interpro
Gene 3D
ProteinModelPortal
S4PKQ2
A0A212FKQ6
A0A2W1BAV0
A0A0N1I581
A0A194R4Y5
A0A0L7R4Y6
+ More
E2B2B9 A0A2A3EFG7 A0A195D177 A0A195B6E3 A0A158NH18 A0A151XDL7 A0A151J205 A0A195FNS9 A0A026VZM3 A0A067RDT9 A0A2A4J394 A0A1B6DKN9 A0A2J7QGK9 A0A310SKW2 A0A1Y1LKG0 K7IRE5 A0A2S2N6L7 A0A2P8XRN2 A0A2S2NW32 A0A2H8TX19 A0A2S2Q3V5 A0A1B6KZ36 A0A232F179 A0A023F1U7 D6WV91 E0VH60 B0WFE1 A0A1Q3EVJ0 U5EZ73 A0A182NU51 A0A084WG14 A0A182J0X5 A0A182L6E4 A0A182RIN9 A0A1S4H1E8 A0A182YDQ3 Q7PVI4 A0A182HGH0 A0A1I8M2W9 A0A182UVZ3 A0A182X0T4 A0A182QH92 Q16TR2 A0A182F3K6 A0A182VUH0 A0A182GRC3 A0A182MG49 A0A0P6K0N0 A0A182K9R6 A0A336LJF4 A0A034VR21 A0A0A1XF30 W5JCR6 A0A182P4D3 A0A0L0C5B3 A0A0P4Y0B1 A0A0K8VVZ4 A0A0K8VQH4 A0A0P5D1P4 A0A0P6GTX1 A0A0P5X730 A0A0P5ACE1 B3N639 E9HKF0 A0A336K4K9 A0A0P6H0P0 B4I1Z4 A0A0J9TP72 B4Q6X9 E2QCR4 Q24009 A0A0P5RDT7 B4NXR7 A0A3B0K8S0 A0A0P5RKI6 A0A0K8UTE3 A0A0K8V9P2 A0A1J1IL71 Q29KC7 A0A1W4WAQ7 B3MKY9 J9K7C8 A0A0M4ERR7 A0A0M4ESA7
E2B2B9 A0A2A3EFG7 A0A195D177 A0A195B6E3 A0A158NH18 A0A151XDL7 A0A151J205 A0A195FNS9 A0A026VZM3 A0A067RDT9 A0A2A4J394 A0A1B6DKN9 A0A2J7QGK9 A0A310SKW2 A0A1Y1LKG0 K7IRE5 A0A2S2N6L7 A0A2P8XRN2 A0A2S2NW32 A0A2H8TX19 A0A2S2Q3V5 A0A1B6KZ36 A0A232F179 A0A023F1U7 D6WV91 E0VH60 B0WFE1 A0A1Q3EVJ0 U5EZ73 A0A182NU51 A0A084WG14 A0A182J0X5 A0A182L6E4 A0A182RIN9 A0A1S4H1E8 A0A182YDQ3 Q7PVI4 A0A182HGH0 A0A1I8M2W9 A0A182UVZ3 A0A182X0T4 A0A182QH92 Q16TR2 A0A182F3K6 A0A182VUH0 A0A182GRC3 A0A182MG49 A0A0P6K0N0 A0A182K9R6 A0A336LJF4 A0A034VR21 A0A0A1XF30 W5JCR6 A0A182P4D3 A0A0L0C5B3 A0A0P4Y0B1 A0A0K8VVZ4 A0A0K8VQH4 A0A0P5D1P4 A0A0P6GTX1 A0A0P5X730 A0A0P5ACE1 B3N639 E9HKF0 A0A336K4K9 A0A0P6H0P0 B4I1Z4 A0A0J9TP72 B4Q6X9 E2QCR4 Q24009 A0A0P5RDT7 B4NXR7 A0A3B0K8S0 A0A0P5RKI6 A0A0K8UTE3 A0A0K8V9P2 A0A1J1IL71 Q29KC7 A0A1W4WAQ7 B3MKY9 J9K7C8 A0A0M4ERR7 A0A0M4ESA7
PDB
6GY4
E-value=8.70404e-22,
Score=257
Ontologies
GO
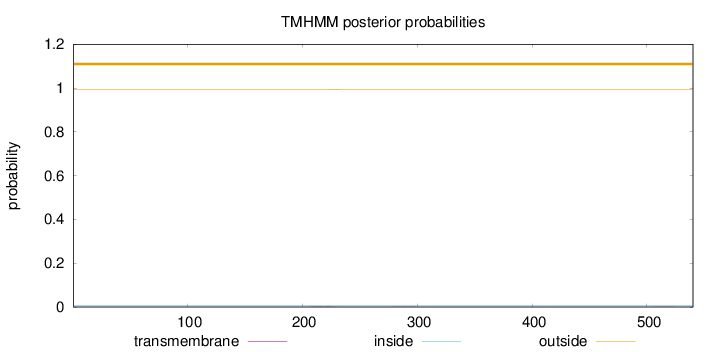
Topology
Length:
540
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03821
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00712
outside
1 - 540
Population Genetic Test Statistics
Pi
226.511417
Theta
180.534407
Tajima's D
0.702208
CLR
0.63798
CSRT
0.573471326433678
Interpretation
Uncertain
