Gene
KWMTBOMO01090
Pre Gene Modal
BGIBMGA007344
Annotation
PREDICTED:_sodium-coupled_neutral_amino_acid_transporter_9-like_[Bombyx_mori]
Full name
Sodium-coupled neutral amino acid transporter 9 homolog
Location in the cell
PlasmaMembrane Reliability : 4.981
Sequence
CDS
ATGCGGTTTCTTAAGAGTTCACCTAGAAAAAGCGAAGATGATCTGTCAACACAGAGCTCTTTGAGGTCAATGGATAGTTGTGAATTATTCGGGAGTGCCTGCTCTGACTATTCTGACTGTGAGTACTGTGATGAGAGAAGACGACAGATTAAAAAAAAGTATTCACCAAGTGACAGCTACTGTAGTGCAATGAGTCGTTTGGAAACGACCCCTTTGCTTGGAGTACTGGCAACAAGACCGGCCTACAAATCCTTGGGAGACAGCACGGATACACTGGGTCCCGGAGAGGTTTCCTCAGATGCCGTATTATCTACTTACAAGAAAAATTTCGAAAAAACTGTCAAAAATGAAAATAAGAAGCAAAGCTCTTTAGTTACAATTTTTTCAATATGGAACACGATAATGGGGTCGTCGCTACTGACCATGGCGTGGGGCGTCGAGCGGGCTGGTCTCCCGGCCGCTCTGGTCCTGGTCGCAGTAATGGCGGCCTTGTGTCTTTACACCGCCTACATATTGATCTCAGTAAACGCTTATCACGGTAGCGACACGTGCTCGGTGCAGGCGCTGTGCCGGCTGCTGCTGGGCGGCTGGGCGGAGGCGCTGGCGCACGTGTTCAGCGTGCTGGTACTCCTCGGAGCGAACATCGTATACTGGATACTCATCACCAACTTCTTTTACTTCACCGTTAATTATTTTATAGATCTAAGTGTGTCGAACGCAACCGAATACAACACGACCCTGCTATGTCCGAATCAAATAGTTAACGAGTCGTTAATGATTCCGAGCCCCGCGGAGGAGTCTCGGTACTGGGGGCTCCACACGACGGCGCCTCTATACACGGCCCTCATTGTCTTCCCGCTCTTAAACTTCAGAAACGTTTCGTTCTTCACTAAATTCAATTCTTTAGGAACCCTATCGGTGTTCTACCTCTTGATCTTCGTAACGGTCAAAGGCTACGCGTGGGGCATCAACATGGGCGCGCTGTGGTCAGAGACCCACGCCGCCAGGAACGCGGCCGTTCTGAGCGGCATGCTGGCCCTATCTTTCTACATCCACAACATCATTATAGAGATCATGAGTAATAACGCAAGGCAAGACAAAAATGGCAGGGATCTAACCATAGCCTTCGTGCTGGTCACGATCACGTACACGATGGTTGGCGCCGTCTTCTACGTCTGCTTCCCGCTCGCCAAGTCCTGCATTGAAGACAACATTCTGAACAACTTCGAGCGTCACGACGTAATGACCGCCGTGGCCAGAGTTCTGTTGCTCTTCCAGGTCGTGACGGTGTACCCGCTGGTGGCGTACATGCTGCGTACGGAGGCGGCCATCTTGTTGTCGTTGCGAGACACGCCCGCCCTTATGTTGCTCGTCAACGTGTGCATAGTTGTCATGTGCATATTGTTCGCGTGCTTCTGCCCTAATATAGGGACAATTATAAGGTACACGGGCGCGCTGAGCGGGCTGGTGCACGTGTTCGCGCTGCCGGCCGCGCTGCACGTGCGCTCGCTGCACCTGCGCGGGGAGCTCGCGCACTGGATGACGTTACTCTACTGTCTCCTCGTGCTCGCCGGGGCCGCCAACCTGCTAATGCAATTCTTCATTACCGAGTGA
Protein
MRFLKSSPRKSEDDLSTQSSLRSMDSCELFGSACSDYSDCEYCDERRRQIKKKYSPSDSYCSAMSRLETTPLLGVLATRPAYKSLGDSTDTLGPGEVSSDAVLSTYKKNFEKTVKNENKKQSSLVTIFSIWNTIMGSSLLTMAWGVERAGLPAALVLVAVMAALCLYTAYILISVNAYHGSDTCSVQALCRLLLGGWAEALAHVFSVLVLLGANIVYWILITNFFYFTVNYFIDLSVSNATEYNTTLLCPNQIVNESLMIPSPAEESRYWGLHTTAPLYTALIVFPLLNFRNVSFFTKFNSLGTLSVFYLLIFVTVKGYAWGINMGALWSETHAARNAAVLSGMLALSFYIHNIIIEIMSNNARQDKNGRDLTIAFVLVTITYTMVGAVFYVCFPLAKSCIEDNILNNFERHDVMTAVARVLLLFQVVTVYPLVAYMLRTEAAILLSLRDTPALMLLVNVCIVVMCILFACFCPNIGTIIRYTGALSGLVHVFALPAALHVRSLHLRGELAHWMTLLYCLLVLAGAANLLMQFFITE
Summary
Description
Lysosomal amino acid transporter involved in the activation of mTORC1 in response to amino acid levels. Probably acts as an amino acid sensor of the Rag GTPases and Ragulator complexes, 2 complexes involved in amino acid sensing and activation of mTORC1, a signaling complex promoting cell growth in response to growth factors, energy levels, and amino acids.
Similarity
Belongs to the amino acid/polyamine transporter 2 family. SLC38A9 subfamily.
Keywords
Amino-acid transport
Complete proteome
Disulfide bond
Endosome
Glycoprotein
Lysosome
Membrane
Metal-binding
Reference proteome
Sodium
Transmembrane
Transmembrane helix
Transport
Feature
chain Sodium-coupled neutral amino acid transporter 9 homolog
Uniprot
A0A2A4ISI3
H9JCU7
A0A2W1BC50
A0A2H1W9C5
A0A0L7KSP1
A0A194RAH9
+ More
A0A212FKR0 A0A1Y1MNC3 J9JJJ9 F4WL87 A0A151I6H5 A0A158NZH8 A0A151WV43 A0A195BPC1 A0A0M9AAM9 A0A2H8TWV6 D2A624 A0A1W4WSP1 A0A026WE26 A0A0A9YEE3 A0A0A9Y6S4 A0A195FCC5 A0A146M953 A0A146L796 A0A146KWI5 A0A2S2PXV9 A0A023F2V8 A0A232FCL7 K7J4T8 E2AB04 A0A2S2PM27 A0A2P8YL72 A0A088ASL6 A0A2J7PXT2 A0A067R137 A0A1B6EGR0 A0A1B6DYX7 A0A1B6IQ56 A0A2J7PXT1 A0A1B6GP34 A0A1B6KXM9 A0A0P5PN38 A0A0N8D107 A0A0P6I656 A0A0P5V4M4 A0A0P5DUB3 A0A0N8DIP9 A0A0N8CRQ7 A0A1I7ZIQ5 A0A0P5MJL0 A0A0P6JCD5 A0A154PH15 A0A2A2L8C9 A0A2A2JQF6 A0A0K0FHV4 A0A0P5Y260 A0A1D2NCA3 A0A0N5BL15 E9GGK7 A0A131YAQ8 A0A087UI50 A0A131YK51 W4XMD1 V5HQ84 A0A224YTD6 A0A1S3KA10 A0A1S3KAJ6 A0A131XPI0 A0A1E1XML5 A0A2G8KUM6 L7M139 A0A1I7S498 A0A131XIN2 A0A0N4ZHU8 A0A090KSG6 A0A0P6EC80 A0A1W2WAC7 A0A0K0EB88 A0A1E1X2Q2 A0A0P4XVN9 F1L1V1 W2TQD0 A0A0P5SCG9 A7SP81 A0A016WK41 A0A016WJY7 A0A0P6CV59 A0A016TGY6 A0A2B4RZW0 A0A293LUP0 A0A061ADY8 Q0G831 A0A016TGV2 Q19425 Q8T3E2 A0A226EN74 A0A1I7UCP6 A0A0N8AIL8 A0A0P5S9L1
A0A212FKR0 A0A1Y1MNC3 J9JJJ9 F4WL87 A0A151I6H5 A0A158NZH8 A0A151WV43 A0A195BPC1 A0A0M9AAM9 A0A2H8TWV6 D2A624 A0A1W4WSP1 A0A026WE26 A0A0A9YEE3 A0A0A9Y6S4 A0A195FCC5 A0A146M953 A0A146L796 A0A146KWI5 A0A2S2PXV9 A0A023F2V8 A0A232FCL7 K7J4T8 E2AB04 A0A2S2PM27 A0A2P8YL72 A0A088ASL6 A0A2J7PXT2 A0A067R137 A0A1B6EGR0 A0A1B6DYX7 A0A1B6IQ56 A0A2J7PXT1 A0A1B6GP34 A0A1B6KXM9 A0A0P5PN38 A0A0N8D107 A0A0P6I656 A0A0P5V4M4 A0A0P5DUB3 A0A0N8DIP9 A0A0N8CRQ7 A0A1I7ZIQ5 A0A0P5MJL0 A0A0P6JCD5 A0A154PH15 A0A2A2L8C9 A0A2A2JQF6 A0A0K0FHV4 A0A0P5Y260 A0A1D2NCA3 A0A0N5BL15 E9GGK7 A0A131YAQ8 A0A087UI50 A0A131YK51 W4XMD1 V5HQ84 A0A224YTD6 A0A1S3KA10 A0A1S3KAJ6 A0A131XPI0 A0A1E1XML5 A0A2G8KUM6 L7M139 A0A1I7S498 A0A131XIN2 A0A0N4ZHU8 A0A090KSG6 A0A0P6EC80 A0A1W2WAC7 A0A0K0EB88 A0A1E1X2Q2 A0A0P4XVN9 F1L1V1 W2TQD0 A0A0P5SCG9 A7SP81 A0A016WK41 A0A016WJY7 A0A0P6CV59 A0A016TGY6 A0A2B4RZW0 A0A293LUP0 A0A061ADY8 Q0G831 A0A016TGV2 Q19425 Q8T3E2 A0A226EN74 A0A1I7UCP6 A0A0N8AIL8 A0A0P5S9L1
Pubmed
19121390
28756777
26227816
26354079
22118469
28004739
+ More
21719571 21347285 18362917 19820115 24508170 30249741 25401762 26823975 25474469 28648823 20075255 20798317 29403074 24845553 26392177 27289101 21292972 26830274 25765539 28797301 29209593 29023486 25576852 21909270 28049606 28503490 21685128 17615350 25730766 9851916 25567906
21719571 21347285 18362917 19820115 24508170 30249741 25401762 26823975 25474469 28648823 20075255 20798317 29403074 24845553 26392177 27289101 21292972 26830274 25765539 28797301 29209593 29023486 25576852 21909270 28049606 28503490 21685128 17615350 25730766 9851916 25567906
EMBL
NWSH01008264
PCG62629.1
BABH01027731
BABH01027732
KZ150177
PZC72558.1
+ More
ODYU01007041 SOQ49442.1 JTDY01006103 KOB66283.1 KQ460685 KPJ12866.1 AGBW02008002 OWR54322.1 GEZM01026177 JAV87173.1 ABLF02023704 GL888207 EGI65050.1 KQ978473 KYM93691.1 ADTU01004623 ADTU01004624 KQ982717 KYQ51724.1 KQ976433 KYM87302.1 KQ435710 KOX79916.1 GFXV01005853 MBW17658.1 KQ971346 EFA05454.2 KK107293 QOIP01000005 EZA53284.1 RLU22491.1 GBHO01015724 JAG27880.1 GBHO01015725 GBRD01015111 JAG27879.1 JAG50715.1 KQ981685 KYN38046.1 GDHC01003322 JAQ15307.1 GDHC01015572 JAQ03057.1 GDHC01019217 JAP99411.1 GGMS01001001 MBY70204.1 GBBI01003338 JAC15374.1 NNAY01000460 OXU28233.1 AAZX01009238 GL438234 EFN69280.1 GGMR01017825 MBY30444.1 PYGN01000517 PSN44983.1 NEVH01020852 PNF21141.1 KK852819 KDR15632.1 GEDC01000181 JAS37117.1 GEDC01006424 JAS30874.1 GECU01018661 JAS89045.1 PNF21142.1 GECZ01005656 JAS64113.1 GEBQ01023764 JAT16213.1 GDIQ01126080 JAL25646.1 GDIP01079391 LRGB01000725 JAM24324.1 KZS16380.1 GDIQ01034946 JAN59791.1 GDIP01105392 JAL98322.1 GDIP01168435 JAJ54967.1 GDIP01029855 JAM73860.1 GDIP01105391 JAL98323.1 GDIQ01155712 JAK96013.1 GDIQ01019477 JAN75260.1 KQ434902 KZC11176.1 LIAE01007070 PAV82297.1 LIAE01010291 PAV63900.1 GDIP01065058 JAM38657.1 LJIJ01000093 ODN02861.1 GL732543 EFX81470.1 GEDV01012580 JAP75977.1 KK119903 KFM77039.1 GEDV01010266 JAP78291.1 AAGJ04062965 AAGJ04062966 GANP01008380 JAB76088.1 GFPF01005956 MAA17102.1 GEFM01006673 JAP69123.1 GFAA01003125 JAU00310.1 MRZV01000359 PIK51716.1 GACK01007477 JAA57557.1 GEFH01003180 JAP65401.1 LN609399 CEF60460.1 GDIQ01070701 JAN24036.1 GFAC01005655 JAT93533.1 GDIP01237144 JAI86257.1 JI169778 ADY44105.1 KI658235 ETN83316.1 GDIP01141553 JAL62161.1 DS469729 EDO34495.1 JARK01000235 EYC39961.1 EYC39960.1 GDIQ01086903 JAN07834.1 JARK01001439 EYC01927.1 LSMT01000190 PFX23994.1 GFWV01006919 MAA31649.1 BX284604 CDR32714.1 CAL36498.1 EYC01926.1 Z68748 CAD27622.1 LNIX01000003 OXA58658.1 GDIP01143793 JAJ79609.1 GDIP01142652 JAL61062.1
ODYU01007041 SOQ49442.1 JTDY01006103 KOB66283.1 KQ460685 KPJ12866.1 AGBW02008002 OWR54322.1 GEZM01026177 JAV87173.1 ABLF02023704 GL888207 EGI65050.1 KQ978473 KYM93691.1 ADTU01004623 ADTU01004624 KQ982717 KYQ51724.1 KQ976433 KYM87302.1 KQ435710 KOX79916.1 GFXV01005853 MBW17658.1 KQ971346 EFA05454.2 KK107293 QOIP01000005 EZA53284.1 RLU22491.1 GBHO01015724 JAG27880.1 GBHO01015725 GBRD01015111 JAG27879.1 JAG50715.1 KQ981685 KYN38046.1 GDHC01003322 JAQ15307.1 GDHC01015572 JAQ03057.1 GDHC01019217 JAP99411.1 GGMS01001001 MBY70204.1 GBBI01003338 JAC15374.1 NNAY01000460 OXU28233.1 AAZX01009238 GL438234 EFN69280.1 GGMR01017825 MBY30444.1 PYGN01000517 PSN44983.1 NEVH01020852 PNF21141.1 KK852819 KDR15632.1 GEDC01000181 JAS37117.1 GEDC01006424 JAS30874.1 GECU01018661 JAS89045.1 PNF21142.1 GECZ01005656 JAS64113.1 GEBQ01023764 JAT16213.1 GDIQ01126080 JAL25646.1 GDIP01079391 LRGB01000725 JAM24324.1 KZS16380.1 GDIQ01034946 JAN59791.1 GDIP01105392 JAL98322.1 GDIP01168435 JAJ54967.1 GDIP01029855 JAM73860.1 GDIP01105391 JAL98323.1 GDIQ01155712 JAK96013.1 GDIQ01019477 JAN75260.1 KQ434902 KZC11176.1 LIAE01007070 PAV82297.1 LIAE01010291 PAV63900.1 GDIP01065058 JAM38657.1 LJIJ01000093 ODN02861.1 GL732543 EFX81470.1 GEDV01012580 JAP75977.1 KK119903 KFM77039.1 GEDV01010266 JAP78291.1 AAGJ04062965 AAGJ04062966 GANP01008380 JAB76088.1 GFPF01005956 MAA17102.1 GEFM01006673 JAP69123.1 GFAA01003125 JAU00310.1 MRZV01000359 PIK51716.1 GACK01007477 JAA57557.1 GEFH01003180 JAP65401.1 LN609399 CEF60460.1 GDIQ01070701 JAN24036.1 GFAC01005655 JAT93533.1 GDIP01237144 JAI86257.1 JI169778 ADY44105.1 KI658235 ETN83316.1 GDIP01141553 JAL62161.1 DS469729 EDO34495.1 JARK01000235 EYC39961.1 EYC39960.1 GDIQ01086903 JAN07834.1 JARK01001439 EYC01927.1 LSMT01000190 PFX23994.1 GFWV01006919 MAA31649.1 BX284604 CDR32714.1 CAL36498.1 EYC01926.1 Z68748 CAD27622.1 LNIX01000003 OXA58658.1 GDIP01143793 JAJ79609.1 GDIP01142652 JAL61062.1
Proteomes
UP000218220
UP000005204
UP000037510
UP000053240
UP000007151
UP000007819
+ More
UP000007755 UP000078542 UP000005205 UP000075809 UP000078540 UP000053105 UP000007266 UP000192223 UP000053097 UP000279307 UP000078541 UP000215335 UP000002358 UP000000311 UP000245037 UP000005203 UP000235965 UP000027135 UP000076858 UP000095287 UP000076502 UP000218231 UP000035680 UP000094527 UP000046392 UP000000305 UP000054359 UP000007110 UP000085678 UP000230750 UP000095284 UP000038045 UP000035681 UP000001593 UP000024635 UP000225706 UP000001940 UP000198287 UP000095282
UP000007755 UP000078542 UP000005205 UP000075809 UP000078540 UP000053105 UP000007266 UP000192223 UP000053097 UP000279307 UP000078541 UP000215335 UP000002358 UP000000311 UP000245037 UP000005203 UP000235965 UP000027135 UP000076858 UP000095287 UP000076502 UP000218231 UP000035680 UP000094527 UP000046392 UP000000305 UP000054359 UP000007110 UP000085678 UP000230750 UP000095284 UP000038045 UP000035681 UP000001593 UP000024635 UP000225706 UP000001940 UP000198287 UP000095282
Interpro
ProteinModelPortal
A0A2A4ISI3
H9JCU7
A0A2W1BC50
A0A2H1W9C5
A0A0L7KSP1
A0A194RAH9
+ More
A0A212FKR0 A0A1Y1MNC3 J9JJJ9 F4WL87 A0A151I6H5 A0A158NZH8 A0A151WV43 A0A195BPC1 A0A0M9AAM9 A0A2H8TWV6 D2A624 A0A1W4WSP1 A0A026WE26 A0A0A9YEE3 A0A0A9Y6S4 A0A195FCC5 A0A146M953 A0A146L796 A0A146KWI5 A0A2S2PXV9 A0A023F2V8 A0A232FCL7 K7J4T8 E2AB04 A0A2S2PM27 A0A2P8YL72 A0A088ASL6 A0A2J7PXT2 A0A067R137 A0A1B6EGR0 A0A1B6DYX7 A0A1B6IQ56 A0A2J7PXT1 A0A1B6GP34 A0A1B6KXM9 A0A0P5PN38 A0A0N8D107 A0A0P6I656 A0A0P5V4M4 A0A0P5DUB3 A0A0N8DIP9 A0A0N8CRQ7 A0A1I7ZIQ5 A0A0P5MJL0 A0A0P6JCD5 A0A154PH15 A0A2A2L8C9 A0A2A2JQF6 A0A0K0FHV4 A0A0P5Y260 A0A1D2NCA3 A0A0N5BL15 E9GGK7 A0A131YAQ8 A0A087UI50 A0A131YK51 W4XMD1 V5HQ84 A0A224YTD6 A0A1S3KA10 A0A1S3KAJ6 A0A131XPI0 A0A1E1XML5 A0A2G8KUM6 L7M139 A0A1I7S498 A0A131XIN2 A0A0N4ZHU8 A0A090KSG6 A0A0P6EC80 A0A1W2WAC7 A0A0K0EB88 A0A1E1X2Q2 A0A0P4XVN9 F1L1V1 W2TQD0 A0A0P5SCG9 A7SP81 A0A016WK41 A0A016WJY7 A0A0P6CV59 A0A016TGY6 A0A2B4RZW0 A0A293LUP0 A0A061ADY8 Q0G831 A0A016TGV2 Q19425 Q8T3E2 A0A226EN74 A0A1I7UCP6 A0A0N8AIL8 A0A0P5S9L1
A0A212FKR0 A0A1Y1MNC3 J9JJJ9 F4WL87 A0A151I6H5 A0A158NZH8 A0A151WV43 A0A195BPC1 A0A0M9AAM9 A0A2H8TWV6 D2A624 A0A1W4WSP1 A0A026WE26 A0A0A9YEE3 A0A0A9Y6S4 A0A195FCC5 A0A146M953 A0A146L796 A0A146KWI5 A0A2S2PXV9 A0A023F2V8 A0A232FCL7 K7J4T8 E2AB04 A0A2S2PM27 A0A2P8YL72 A0A088ASL6 A0A2J7PXT2 A0A067R137 A0A1B6EGR0 A0A1B6DYX7 A0A1B6IQ56 A0A2J7PXT1 A0A1B6GP34 A0A1B6KXM9 A0A0P5PN38 A0A0N8D107 A0A0P6I656 A0A0P5V4M4 A0A0P5DUB3 A0A0N8DIP9 A0A0N8CRQ7 A0A1I7ZIQ5 A0A0P5MJL0 A0A0P6JCD5 A0A154PH15 A0A2A2L8C9 A0A2A2JQF6 A0A0K0FHV4 A0A0P5Y260 A0A1D2NCA3 A0A0N5BL15 E9GGK7 A0A131YAQ8 A0A087UI50 A0A131YK51 W4XMD1 V5HQ84 A0A224YTD6 A0A1S3KA10 A0A1S3KAJ6 A0A131XPI0 A0A1E1XML5 A0A2G8KUM6 L7M139 A0A1I7S498 A0A131XIN2 A0A0N4ZHU8 A0A090KSG6 A0A0P6EC80 A0A1W2WAC7 A0A0K0EB88 A0A1E1X2Q2 A0A0P4XVN9 F1L1V1 W2TQD0 A0A0P5SCG9 A7SP81 A0A016WK41 A0A016WJY7 A0A0P6CV59 A0A016TGY6 A0A2B4RZW0 A0A293LUP0 A0A061ADY8 Q0G831 A0A016TGV2 Q19425 Q8T3E2 A0A226EN74 A0A1I7UCP6 A0A0N8AIL8 A0A0P5S9L1
PDB
6C08
E-value=1.77467e-72,
Score=694
Ontologies
GO
PANTHER
Topology
Subcellular location
Lysosome membrane
Late endosome membrane
Late endosome membrane
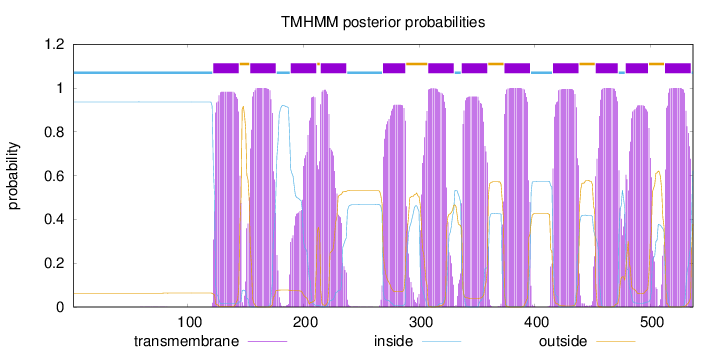
Length:
537
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
247.29173
Exp number, first 60 AAs:
0.00083
Total prob of N-in:
0.93669
inside
1 - 121
TMhelix
122 - 144
outside
145 - 153
TMhelix
154 - 176
inside
177 - 188
TMhelix
189 - 211
outside
212 - 214
TMhelix
215 - 237
inside
238 - 268
TMhelix
269 - 288
outside
289 - 307
TMhelix
308 - 330
inside
331 - 336
TMhelix
337 - 359
outside
360 - 373
TMhelix
374 - 396
inside
397 - 415
TMhelix
416 - 438
outside
439 - 452
TMhelix
453 - 472
inside
473 - 478
TMhelix
479 - 498
outside
499 - 512
TMhelix
513 - 535
inside
536 - 537
Population Genetic Test Statistics
Pi
242.474109
Theta
156.41281
Tajima's D
2.025118
CLR
0.356349
CSRT
0.887605619719014
Interpretation
Uncertain
