Gene
KWMTBOMO01089 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007425
Annotation
PREDICTED:_phosphoglucomutase_[Amyelois_transitella]
Full name
Phosphoglucomutase
Alternative Name
Glucose phosphomutase
Location in the cell
Cytoplasmic Reliability : 2.147
Sequence
CDS
ATGTCTAACAGTGTAACGGTTGACACTAAACCGTATGAGGGCCAAAAGCCGGGCACAAGTGGATTGCGTAAAAAAGTTAAAGTTTTCGTTCAAGAAAACTACACGGAGAATTTTATTCAATGCATTTTGGACGTGAATAAAACCTCTTTGATAGGATCGACGCTGGTTGTCGGAGGAGACGGAAGGTATCTCGTGAAAGAGGTTGTTGATAAAATTATTAAGATCAGTGCAGCGAATGGGGTTTCGAAATTGATAGTAGGACAAAATGGAATACTATCAACTCCTGCTGTGTCTTATATTATTAGAAAATATAAAACTCTCGGTGGTATAGTTCTCACGGCGTCCCACAATCCGGGCGGTGTGGACGAAGACTTTGGCATAAAGTTCAACTGCAGTAACGGAGGCCCAGCTTCCGATGCTACCACCGATGCCATCTACAAACTCACCACCTCGATAAAGCAGTACAAGATTGTTCCAGATTTAAACTGCGCCATTGACAAAATCGACGTACATACTTTTCAGGTAGATCAGCGGCAGTTTACAGTCGAAGTCATAGATGCAGTGGAAGATTATGTTTCGTACATGAAGGAGATCTTCGATTTCCCGAAGATCAAGGCTTTGATCCAGGGAACGGAGCAGAGGAAGCCCTTCAACGTGCTCATTGATGCTATGAACGGAGTAACCGGACCCTACGTGAAGCGGATATTCTTAGAGGAGCTAGGTTGTTCCGAGAAGAACGTGCGACGTATCGTGCCCCTGGAAGACTTCGGCGGGGCTCATCCGGATCCGAACTTGACGTACGCCGCGGATTTGGTCGAGGCTGTCAAAGAAGGGGACTATGATTTCGGGGCCGCTTTCGACGGAGACGGGGACCGCAACATGATACTAGGGCGGGACGCGTTCTTCGTGACGCCGTCGGACTCCCTGGCGGTGCTGGCCGACAATCTGGAGTGCATACCGTACTTCGCGCGAGGGGGCGTGCGGGGGCTGGCGCGCAGCATGCCCACCGCCGCCGCCGTGGATCGGGTGGCGGCCGCGCGCGCCCTGCCCCTCTTCGAGGTGCCCACCGGTTGGAAATACTTTGGCAACCTGATGGACGCGGGCCAGCTGTCGCTGTGCGGAGAGGAGAGCTTCGGTACCGGGTCCGACCACGTGCGGGAGAAGGACGGGCTGTGGGCGGCGCTGGCCTGGCTCTCCCTCCTGGCGCACTCCGGCCGGTCCGTGCAGGAGATGCTGCAGGCGCACTGGAAACAGTTCGGGAGGAACTACTTCACGAGGTACGACTACGAGAACTGTTCCTCGGAGTCCTGCGAGGAGATGATGAGCGTCCTTGAGCGCACCATGACCTCGCCCGGGTTCGTGGGCTCGGCGCACTCCGCCGGCGGTAAGGAGTACGTCGTCAAGCTAGCCGACAACTTCTCCTACGTGGACCCCATCGATCAGAGCGTGGCCAAGAGACAGGGCCTGCGGATAATATTCGAGGACGGATCCCGCATAGTGTTCCGTCTCAGCGGCACGGGCAGTTCCGGCGCTACCGTCAGAGTCTACGTGGACTCGTACGAGGCGGTGAGCGTGGGGGGCGCCGCTGCTGACGCGCTGGGGCCGCTCGTCAGCATCGCCCTGGACATATCCCGCTTGAAGCAATACACGGGTCGGGACGAGCCCACAGTCATCACTTAA
Protein
MSNSVTVDTKPYEGQKPGTSGLRKKVKVFVQENYTENFIQCILDVNKTSLIGSTLVVGGDGRYLVKEVVDKIIKISAANGVSKLIVGQNGILSTPAVSYIIRKYKTLGGIVLTASHNPGGVDEDFGIKFNCSNGGPASDATTDAIYKLTTSIKQYKIVPDLNCAIDKIDVHTFQVDQRQFTVEVIDAVEDYVSYMKEIFDFPKIKALIQGTEQRKPFNVLIDAMNGVTGPYVKRIFLEELGCSEKNVRRIVPLEDFGGAHPDPNLTYAADLVEAVKEGDYDFGAAFDGDGDRNMILGRDAFFVTPSDSLAVLADNLECIPYFARGGVRGLARSMPTAAAVDRVAAARALPLFEVPTGWKYFGNLMDAGQLSLCGEESFGTGSDHVREKDGLWAALAWLSLLAHSGRSVQEMLQAHWKQFGRNYFTRYDYENCSSESCEEMMSVLERTMTSPGFVGSAHSAGGKEYVVKLADNFSYVDPIDQSVAKRQGLRIIFEDGSRIVFRLSGTGSSGATVRVYVDSYEAVSVGGAAADALGPLVSIALDISRLKQYTGRDEPTVIT
Summary
Description
This enzyme participates in both the breakdown and synthesis of glucose.
Catalytic Activity
alpha-D-glucose 1-phosphate = alpha-D-glucose 6-phosphate
Cofactor
Mg(2+)
Biophysicochemical Properties
140 uM for glucose-1-phosphate (for Pgm-A at pH 6.0)
158 uM for glucose-1-phosphate (for Pgm-A at pH 7.4)
4.4 uM for glucose-1,6-diphosphate (for Pgm-A at pH 6.0)
4.4 uM for glucose-1,6-diphosphate (for Pgm-A at pH 7.4)
142.2 uM for glucose-1-phosphate (for Pgm-B at pH 6.0)
112.4 uM for glucose-1-phosphate (for Pgm-B at pH 7.4)
21.7 uM for glucose-1,6-diphosphate (for Pgm-B at pH 6.0)
4.2 uM for glucose-1,6-diphosphate (for Pgm-B at pH 7.4)
158 uM for glucose-1-phosphate (for Pgm-A at pH 7.4)
4.4 uM for glucose-1,6-diphosphate (for Pgm-A at pH 6.0)
4.4 uM for glucose-1,6-diphosphate (for Pgm-A at pH 7.4)
142.2 uM for glucose-1-phosphate (for Pgm-B at pH 6.0)
112.4 uM for glucose-1-phosphate (for Pgm-B at pH 7.4)
21.7 uM for glucose-1,6-diphosphate (for Pgm-B at pH 6.0)
4.2 uM for glucose-1,6-diphosphate (for Pgm-B at pH 7.4)
Similarity
Belongs to the phosphohexose mutase family.
Belongs to the adaptor complexes medium subunit family.
Belongs to the adaptor complexes medium subunit family.
Keywords
Carbohydrate metabolism
Glucose metabolism
Isomerase
Magnesium
Metal-binding
Phosphoprotein
Polymorphism
Complete proteome
Reference proteome
Feature
chain Phosphoglucomutase
Uniprot
H9JD28
A0A2W1BH83
S4NNG7
A0A2H1W8N8
E1U7I4
A0A194R6J8
+ More
A0A0N1I3K6 A0A1B6JU52 A0A0K8UNQ3 A0A1B6C217 W8BDR3 A0A131Y3J0 B4MM44 A0A147BV33 A0A034VEB3 A0A1Y1LPW3 B0WEU6 A0A154PH97 A0A0J9RWA8 A0A1Q3FKJ2 Q7KHA1 A0A3B0KME6 A0A1W4UGN4 U5ELU3 Q9GQ67 B4PI34 B3NI51 A0A2A4ITA8 Q9VUY9 A0A1I8NQ07 A0A1I8NQ09 Q29EN2 A0A336MMP4 A0A1S4FP29 A0A3B0JY52 Q16U43 A0A1W4WSY5 B4H296 B4LDE2 T1PCB7 A0A182G2L5 A0A182Q403 E9J743 J9JMZ9 A0A1I8M689 V5I8J8 A0A2M4A6T3 A0A026WBF7 A0A1A9W7M1 A0A182YEF1 Q58I84 B3M958 A0A182JRR5 A0A0M9A9D5 A0A088ASJ9 A0A182KPX6 A0A182XMC2 A0A2M3YZ53 A0A182U8R6 A0A182V3C5 A0A0P5XC22 A0A182I257 A0A182PTN1 A0A0P5FDA1 A0A164ZBS7 A0A2M4BJ32 Q7Q6G4 A0A084WEB1 B4L9P4 A0A182MMX7 A0A3L8DQF8 E9GBC4 A0A182NJD6 A0A182FH19 N6TII1 W5J6Z3 A0A0P5T336 A0A182R4I5 A0A346A6M0 D3TM66 J3JVE4 A0A182ITM5 A0A182WLC1 A0A0P4VPJ3 A0A0M4EYD5 A0A2J7PXV5 A0A1L8E1W5 T1E9Z7 B4IZY5 A0A1A9ZF25 A0A1L8E216 A0A1S3KAE3 A0A2A3EEZ1 A0A232FCN4 K7IN84 A0A023EWH2 A0A1J1I236 A0A2P6L0L9 A0A151I722 C0KJJ8
A0A0N1I3K6 A0A1B6JU52 A0A0K8UNQ3 A0A1B6C217 W8BDR3 A0A131Y3J0 B4MM44 A0A147BV33 A0A034VEB3 A0A1Y1LPW3 B0WEU6 A0A154PH97 A0A0J9RWA8 A0A1Q3FKJ2 Q7KHA1 A0A3B0KME6 A0A1W4UGN4 U5ELU3 Q9GQ67 B4PI34 B3NI51 A0A2A4ITA8 Q9VUY9 A0A1I8NQ07 A0A1I8NQ09 Q29EN2 A0A336MMP4 A0A1S4FP29 A0A3B0JY52 Q16U43 A0A1W4WSY5 B4H296 B4LDE2 T1PCB7 A0A182G2L5 A0A182Q403 E9J743 J9JMZ9 A0A1I8M689 V5I8J8 A0A2M4A6T3 A0A026WBF7 A0A1A9W7M1 A0A182YEF1 Q58I84 B3M958 A0A182JRR5 A0A0M9A9D5 A0A088ASJ9 A0A182KPX6 A0A182XMC2 A0A2M3YZ53 A0A182U8R6 A0A182V3C5 A0A0P5XC22 A0A182I257 A0A182PTN1 A0A0P5FDA1 A0A164ZBS7 A0A2M4BJ32 Q7Q6G4 A0A084WEB1 B4L9P4 A0A182MMX7 A0A3L8DQF8 E9GBC4 A0A182NJD6 A0A182FH19 N6TII1 W5J6Z3 A0A0P5T336 A0A182R4I5 A0A346A6M0 D3TM66 J3JVE4 A0A182ITM5 A0A182WLC1 A0A0P4VPJ3 A0A0M4EYD5 A0A2J7PXV5 A0A1L8E1W5 T1E9Z7 B4IZY5 A0A1A9ZF25 A0A1L8E216 A0A1S3KAE3 A0A2A3EEZ1 A0A232FCN4 K7IN84 A0A023EWH2 A0A1J1I236 A0A2P6L0L9 A0A151I722 C0KJJ8
EC Number
5.4.2.2
Pubmed
19121390
28756777
23622113
26354079
24495485
17994087
+ More
29652888 25348373 28004739 22936249 11102370 17550304 12049662 10731132 12537572 44190 18327897 11290720 11560897 15632085 17510324 26483478 21282665 25315136 24508170 25244985 20966253 12364791 14747013 17210077 24438588 30249741 21292972 23537049 20920257 23761445 29187976 20353571 22516182 28648823 20075255 24945155
29652888 25348373 28004739 22936249 11102370 17550304 12049662 10731132 12537572 44190 18327897 11290720 11560897 15632085 17510324 26483478 21282665 25315136 24508170 25244985 20966253 12364791 14747013 17210077 24438588 30249741 21292972 23537049 20920257 23761445 29187976 20353571 22516182 28648823 20075255 24945155
EMBL
BABH01027730
KZ150177
PZC72557.1
GAIX01013936
JAA78624.1
ODYU01007041
+ More
SOQ49441.1 FJ793041 ACY69180.1 KQ460685 KPJ12865.1 KQ459072 KPJ03853.1 GECU01005000 JAT02707.1 GDHF01024005 JAI28309.1 GEDC01029894 JAS07404.1 GAMC01011429 GAMC01011427 JAB95128.1 GEFM01002718 JAP73078.1 CH963847 EDW73053.1 GEGO01000824 JAR94580.1 GAKP01018507 GAKP01018505 JAC40447.1 GEZM01050186 JAV75683.1 DS231911 EDS45859.1 KQ434902 KZC11177.1 CM002912 KMY99951.1 GFDL01007021 JAV28024.1 AF290357 AF290358 AF290359 AF290360 AF290361 AF290362 AF290363 AF290364 AF290365 AF290366 AF290367 AF290368 AF290369 AAG42290.1 AAG42291.1 AAG42292.1 AAG42293.1 AAG42294.1 AAG42295.1 AAG42296.1 AAG42297.1 AAG42298.1 AAG42299.1 AAG42300.1 AAG42301.1 AAG42302.1 OUUW01000012 SPP86996.1 GANO01001270 JAB58601.1 AF290370 AAG42303.1 CM000159 EDW95482.1 CH954178 EDV52207.1 NWSH01008264 PCG62628.1 AF290313 AF290314 AF290315 AF290316 AF290317 AF290318 AF290319 AF290320 AF290321 AF290322 AF290323 AF290324 AF290325 AF290326 AF290327 AF290328 AF290329 AF290330 AF290331 AF290332 AF290333 AF290334 AF290335 AF290336 AF290337 AF290338 AF290339 AF290340 AF290341 AF290342 AF290343 AF290344 AF290345 AF290346 AF290347 AF290348 AF290349 AF290350 AF290351 AF290352 AF290353 AF290354 AF290355 AF290356 AF416981 AF416982 AF416983 AF416984 AE014296 BT010043 AAF49533.1 AAG44900.1 AAG44901.1 AAG44902.1 AAG44903.1 AAG44904.1 AAG44905.1 AAG44906.1 AAG44907.1 AAG44908.1 AAG44909.1 AAG44910.1 AAG44911.1 AAG44912.1 AAG44913.1 AAG44914.1 AAG44915.1 AAG44916.1 AAG44917.1 AAG44918.1 AAG44919.1 AAG44920.1 AAG44921.1 AAG44922.1 AAG44923.1 AAG44924.1 AAG44925.1 AAG44926.1 AAG44927.1 AAG44928.1 AAG44929.1 AAG44930.1 AAG44931.1 AAG44932.1 AAG44933.1 AAG44934.1 AAG44935.1 AAG44936.1 AAG44937.1 AAG44938.1 AAG44939.1 AAG44940.1 AAG44941.1 AAG44942.1 AAG44943.1 AAL08565.1 AAL08568.1 AAQ22512.1 CH379070 EAL30028.1 UFQS01001397 UFQT01001397 SSX10687.1 SSX30369.1 SPP86997.1 CH477632 EAT38042.1 CH479203 EDW30448.1 CH940647 EDW68880.1 KA645583 AFP60212.1 JXUM01006654 KQ560219 KXJ83762.1 AXCN02000448 GL768421 EFZ11363.1 ABLF02017858 ABLF02023700 ABLF02023704 GALX01004560 JAB63906.1 GGFK01003131 MBW36452.1 KK107293 EZA53283.1 AY944755 AAX47078.1 CH902618 EDV40042.1 KQ435710 KOX79915.1 GGFM01000799 MBW21550.1 GDIP01074252 JAM29463.1 APCN01000123 GDIQ01257217 GDIQ01054612 JAJ94507.1 LRGB01000745 KZS16178.1 GGFJ01003901 MBW53042.1 AAAB01008960 EAA11635.2 ATLV01023198 KE525341 KFB48555.1 CH933816 EDW17092.1 AXCM01002346 QOIP01000005 RLU22492.1 GL732538 EFX83182.1 APGK01036673 KB740941 KB632225 ENN77623.1 ERL90249.1 ADMH02002052 ETN59746.1 GDIQ01097040 JAL54686.1 MF908520 AXK92642.1 EZ422518 ADD18794.1 BT127212 AEE62174.1 GDRN01108558 JAI57209.1 CP012525 ALC43182.1 NEVH01020852 PNF21144.1 GFDF01001387 JAV12697.1 GAMD01001468 JAB00123.1 CH916366 EDV96757.1 GFDF01001388 JAV12696.1 KZ288271 PBC29769.1 NNAY01000480 OXU28087.1 GAPW01000774 JAC12824.1 CVRI01000035 CRK92902.1 MWRG01002894 PRD32122.1 KQ978473 KYM93692.1 FJ609649 ACM78949.1
SOQ49441.1 FJ793041 ACY69180.1 KQ460685 KPJ12865.1 KQ459072 KPJ03853.1 GECU01005000 JAT02707.1 GDHF01024005 JAI28309.1 GEDC01029894 JAS07404.1 GAMC01011429 GAMC01011427 JAB95128.1 GEFM01002718 JAP73078.1 CH963847 EDW73053.1 GEGO01000824 JAR94580.1 GAKP01018507 GAKP01018505 JAC40447.1 GEZM01050186 JAV75683.1 DS231911 EDS45859.1 KQ434902 KZC11177.1 CM002912 KMY99951.1 GFDL01007021 JAV28024.1 AF290357 AF290358 AF290359 AF290360 AF290361 AF290362 AF290363 AF290364 AF290365 AF290366 AF290367 AF290368 AF290369 AAG42290.1 AAG42291.1 AAG42292.1 AAG42293.1 AAG42294.1 AAG42295.1 AAG42296.1 AAG42297.1 AAG42298.1 AAG42299.1 AAG42300.1 AAG42301.1 AAG42302.1 OUUW01000012 SPP86996.1 GANO01001270 JAB58601.1 AF290370 AAG42303.1 CM000159 EDW95482.1 CH954178 EDV52207.1 NWSH01008264 PCG62628.1 AF290313 AF290314 AF290315 AF290316 AF290317 AF290318 AF290319 AF290320 AF290321 AF290322 AF290323 AF290324 AF290325 AF290326 AF290327 AF290328 AF290329 AF290330 AF290331 AF290332 AF290333 AF290334 AF290335 AF290336 AF290337 AF290338 AF290339 AF290340 AF290341 AF290342 AF290343 AF290344 AF290345 AF290346 AF290347 AF290348 AF290349 AF290350 AF290351 AF290352 AF290353 AF290354 AF290355 AF290356 AF416981 AF416982 AF416983 AF416984 AE014296 BT010043 AAF49533.1 AAG44900.1 AAG44901.1 AAG44902.1 AAG44903.1 AAG44904.1 AAG44905.1 AAG44906.1 AAG44907.1 AAG44908.1 AAG44909.1 AAG44910.1 AAG44911.1 AAG44912.1 AAG44913.1 AAG44914.1 AAG44915.1 AAG44916.1 AAG44917.1 AAG44918.1 AAG44919.1 AAG44920.1 AAG44921.1 AAG44922.1 AAG44923.1 AAG44924.1 AAG44925.1 AAG44926.1 AAG44927.1 AAG44928.1 AAG44929.1 AAG44930.1 AAG44931.1 AAG44932.1 AAG44933.1 AAG44934.1 AAG44935.1 AAG44936.1 AAG44937.1 AAG44938.1 AAG44939.1 AAG44940.1 AAG44941.1 AAG44942.1 AAG44943.1 AAL08565.1 AAL08568.1 AAQ22512.1 CH379070 EAL30028.1 UFQS01001397 UFQT01001397 SSX10687.1 SSX30369.1 SPP86997.1 CH477632 EAT38042.1 CH479203 EDW30448.1 CH940647 EDW68880.1 KA645583 AFP60212.1 JXUM01006654 KQ560219 KXJ83762.1 AXCN02000448 GL768421 EFZ11363.1 ABLF02017858 ABLF02023700 ABLF02023704 GALX01004560 JAB63906.1 GGFK01003131 MBW36452.1 KK107293 EZA53283.1 AY944755 AAX47078.1 CH902618 EDV40042.1 KQ435710 KOX79915.1 GGFM01000799 MBW21550.1 GDIP01074252 JAM29463.1 APCN01000123 GDIQ01257217 GDIQ01054612 JAJ94507.1 LRGB01000745 KZS16178.1 GGFJ01003901 MBW53042.1 AAAB01008960 EAA11635.2 ATLV01023198 KE525341 KFB48555.1 CH933816 EDW17092.1 AXCM01002346 QOIP01000005 RLU22492.1 GL732538 EFX83182.1 APGK01036673 KB740941 KB632225 ENN77623.1 ERL90249.1 ADMH02002052 ETN59746.1 GDIQ01097040 JAL54686.1 MF908520 AXK92642.1 EZ422518 ADD18794.1 BT127212 AEE62174.1 GDRN01108558 JAI57209.1 CP012525 ALC43182.1 NEVH01020852 PNF21144.1 GFDF01001387 JAV12697.1 GAMD01001468 JAB00123.1 CH916366 EDV96757.1 GFDF01001388 JAV12696.1 KZ288271 PBC29769.1 NNAY01000480 OXU28087.1 GAPW01000774 JAC12824.1 CVRI01000035 CRK92902.1 MWRG01002894 PRD32122.1 KQ978473 KYM93692.1 FJ609649 ACM78949.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007798
UP000002320
UP000076502
+ More
UP000268350 UP000192221 UP000002282 UP000008711 UP000218220 UP000000803 UP000095300 UP000001819 UP000008820 UP000192223 UP000008744 UP000008792 UP000069940 UP000249989 UP000075886 UP000007819 UP000095301 UP000053097 UP000091820 UP000076408 UP000007801 UP000075881 UP000053105 UP000005203 UP000075882 UP000076407 UP000075902 UP000075903 UP000075840 UP000075885 UP000076858 UP000007062 UP000030765 UP000009192 UP000075883 UP000279307 UP000000305 UP000075884 UP000069272 UP000019118 UP000030742 UP000000673 UP000075900 UP000075880 UP000075920 UP000092553 UP000235965 UP000001070 UP000092445 UP000085678 UP000242457 UP000215335 UP000002358 UP000183832 UP000078542
UP000268350 UP000192221 UP000002282 UP000008711 UP000218220 UP000000803 UP000095300 UP000001819 UP000008820 UP000192223 UP000008744 UP000008792 UP000069940 UP000249989 UP000075886 UP000007819 UP000095301 UP000053097 UP000091820 UP000076408 UP000007801 UP000075881 UP000053105 UP000005203 UP000075882 UP000076407 UP000075902 UP000075903 UP000075840 UP000075885 UP000076858 UP000007062 UP000030765 UP000009192 UP000075883 UP000279307 UP000000305 UP000075884 UP000069272 UP000019118 UP000030742 UP000000673 UP000075900 UP000075880 UP000075920 UP000092553 UP000235965 UP000001070 UP000092445 UP000085678 UP000242457 UP000215335 UP000002358 UP000183832 UP000078542
PRIDE
Pfam
Interpro
IPR005846
A-D-PHexomutase_a/b/a-III
+ More
IPR016066 A-D-PHexomutase_CS
IPR036900 A-D-PHexomutase_C_sf
IPR005845 A-D-PHexomutase_a/b/a-II
IPR005844 A-D-PHexomutase_a/b/a-I
IPR005841 Alpha-D-phosphohexomutase_SF
IPR005843 A-D-PHexomutase_C
IPR016055 A-D-PHexomutase_a/b/a-I/II/III
IPR011012 Longin-like_dom_sf
IPR028565 MHD
IPR022775 AP_mu_sigma_su
IPR036168 AP2_Mu_C_sf
IPR016066 A-D-PHexomutase_CS
IPR036900 A-D-PHexomutase_C_sf
IPR005845 A-D-PHexomutase_a/b/a-II
IPR005844 A-D-PHexomutase_a/b/a-I
IPR005841 Alpha-D-phosphohexomutase_SF
IPR005843 A-D-PHexomutase_C
IPR016055 A-D-PHexomutase_a/b/a-I/II/III
IPR011012 Longin-like_dom_sf
IPR028565 MHD
IPR022775 AP_mu_sigma_su
IPR036168 AP2_Mu_C_sf
ProteinModelPortal
H9JD28
A0A2W1BH83
S4NNG7
A0A2H1W8N8
E1U7I4
A0A194R6J8
+ More
A0A0N1I3K6 A0A1B6JU52 A0A0K8UNQ3 A0A1B6C217 W8BDR3 A0A131Y3J0 B4MM44 A0A147BV33 A0A034VEB3 A0A1Y1LPW3 B0WEU6 A0A154PH97 A0A0J9RWA8 A0A1Q3FKJ2 Q7KHA1 A0A3B0KME6 A0A1W4UGN4 U5ELU3 Q9GQ67 B4PI34 B3NI51 A0A2A4ITA8 Q9VUY9 A0A1I8NQ07 A0A1I8NQ09 Q29EN2 A0A336MMP4 A0A1S4FP29 A0A3B0JY52 Q16U43 A0A1W4WSY5 B4H296 B4LDE2 T1PCB7 A0A182G2L5 A0A182Q403 E9J743 J9JMZ9 A0A1I8M689 V5I8J8 A0A2M4A6T3 A0A026WBF7 A0A1A9W7M1 A0A182YEF1 Q58I84 B3M958 A0A182JRR5 A0A0M9A9D5 A0A088ASJ9 A0A182KPX6 A0A182XMC2 A0A2M3YZ53 A0A182U8R6 A0A182V3C5 A0A0P5XC22 A0A182I257 A0A182PTN1 A0A0P5FDA1 A0A164ZBS7 A0A2M4BJ32 Q7Q6G4 A0A084WEB1 B4L9P4 A0A182MMX7 A0A3L8DQF8 E9GBC4 A0A182NJD6 A0A182FH19 N6TII1 W5J6Z3 A0A0P5T336 A0A182R4I5 A0A346A6M0 D3TM66 J3JVE4 A0A182ITM5 A0A182WLC1 A0A0P4VPJ3 A0A0M4EYD5 A0A2J7PXV5 A0A1L8E1W5 T1E9Z7 B4IZY5 A0A1A9ZF25 A0A1L8E216 A0A1S3KAE3 A0A2A3EEZ1 A0A232FCN4 K7IN84 A0A023EWH2 A0A1J1I236 A0A2P6L0L9 A0A151I722 C0KJJ8
A0A0N1I3K6 A0A1B6JU52 A0A0K8UNQ3 A0A1B6C217 W8BDR3 A0A131Y3J0 B4MM44 A0A147BV33 A0A034VEB3 A0A1Y1LPW3 B0WEU6 A0A154PH97 A0A0J9RWA8 A0A1Q3FKJ2 Q7KHA1 A0A3B0KME6 A0A1W4UGN4 U5ELU3 Q9GQ67 B4PI34 B3NI51 A0A2A4ITA8 Q9VUY9 A0A1I8NQ07 A0A1I8NQ09 Q29EN2 A0A336MMP4 A0A1S4FP29 A0A3B0JY52 Q16U43 A0A1W4WSY5 B4H296 B4LDE2 T1PCB7 A0A182G2L5 A0A182Q403 E9J743 J9JMZ9 A0A1I8M689 V5I8J8 A0A2M4A6T3 A0A026WBF7 A0A1A9W7M1 A0A182YEF1 Q58I84 B3M958 A0A182JRR5 A0A0M9A9D5 A0A088ASJ9 A0A182KPX6 A0A182XMC2 A0A2M3YZ53 A0A182U8R6 A0A182V3C5 A0A0P5XC22 A0A182I257 A0A182PTN1 A0A0P5FDA1 A0A164ZBS7 A0A2M4BJ32 Q7Q6G4 A0A084WEB1 B4L9P4 A0A182MMX7 A0A3L8DQF8 E9GBC4 A0A182NJD6 A0A182FH19 N6TII1 W5J6Z3 A0A0P5T336 A0A182R4I5 A0A346A6M0 D3TM66 J3JVE4 A0A182ITM5 A0A182WLC1 A0A0P4VPJ3 A0A0M4EYD5 A0A2J7PXV5 A0A1L8E1W5 T1E9Z7 B4IZY5 A0A1A9ZF25 A0A1L8E216 A0A1S3KAE3 A0A2A3EEZ1 A0A232FCN4 K7IN84 A0A023EWH2 A0A1J1I236 A0A2P6L0L9 A0A151I722 C0KJJ8
PDB
3PMG
E-value=5.98662e-168,
Score=1518
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00230 Purine metabolism - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00230 Purine metabolism - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
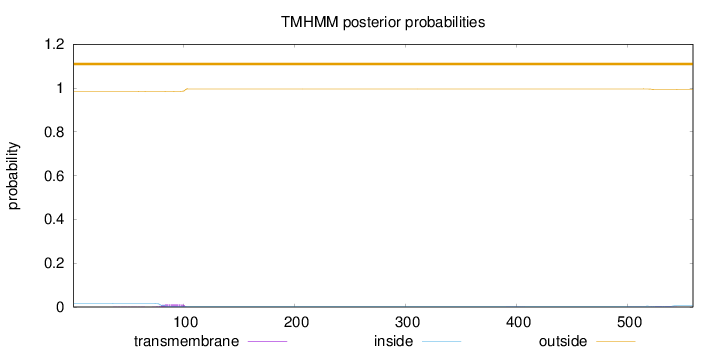
Length:
559
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.32987
Exp number, first 60 AAs:
0.00735
Total prob of N-in:
0.01642
outside
1 - 559
Population Genetic Test Statistics
Pi
263.10696
Theta
161.461531
Tajima's D
2.063753
CLR
0.399103
CSRT
0.893105344732763
Interpretation
Uncertain
