Pre Gene Modal
BGIBMGA007345
Annotation
PREDICTED:_fatty-acid_amide_hydrolase_2-like_isoform_X1_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.898
Sequence
CDS
ATGTGCACAACAAACCACGGCGACGGAAGAATTTCGCATTCTGTGCCTGGCAGCAAAACTTGTAACATTATTCGAGGAATAGCATTTAACATGATCAAACACGCATTCCTGGTCTTCAGAACATATCTGGACTTATTCATTGATTACATATTCTCCCTTTACTGGGAAGCGAAGAGGACACCTTTGCCAAATTTGGAAAAAAAGCATATTATACTTAAGGACGATGCTGTGACCATTGCCGCAAAGATCAGAAATAGACAGTTAAAGTCTGAAGATTTGGTTCAAATGTGCATTGAAAGAATAAAAATTGTCAATCCAATACTGAACGCCGTCACAGATGAGCGTTTTGAAGAGGCTTTAGAAGAAGCGCGCGAAATCGACCGGATGCTTGATGAGGGACTCTCAGAAGAATATCTCAAGAAGAAACCATTCCTTGGTGTTCCGTTTACGGCGAAAGAGAGTCACGCGGTGAAGGGCCTGCTCCACACGTTGGGCATCAAGGCGCGGGCGCACGTCCGCGCGCACGAGGACGCGGAGTGCGTGCGCCTCATGCGCGCGGCCGGCGCCCTCCCGCTCGCCGTCACCAACGTACCAGAGATCAACAAATGGTGTGAAACTCGCAACATGGTGTTTGGTCAGACCAACAACCCGTATCACACCGGGCGCACCACGGGCGGGTCCAGCGGTGGGGAGGCCGCGCTGATGGCCGCCATGGCGACACCCATATCGTTATGTTCGGACATCGGCGGGTCCACGCGGATGCCGGCGTTCTACTGCGGCTTGTTCGGCATCAACCCCACCGCCGGATATACCAGTCTTAAAGGCTCGGCGCTGCGCTCCGGTAAAGATGACACCATGGCGTCGATAGGGTTCGTTAGCAGACACACCTGCGACCTCGCCGCGCTCACCAAAGTCATCCTGGCCGACAAGGCTGCTGAGCTGGACTTGGACAGGCCCGTGGACCTCAAGACGATCAAGTATTATTACCTGGACACGGCCCAGGACCTGCGTGTCAGTCCCATCCAGCCGGAGCTCCGGGAGGCCATGGACAAAGTCATAAGTCAACTAAGCGAGGAGGCGTCGAGCGCCCGGCGCTACCAGCACGCCGGCCTCGACCACATGTACGCGCTGTGGCGGCACGCCATGACCAGGGAGGCCGACGTGTTCGGGCGCCTCCTCACCAACAACCAAGGAGAGGCCAGCGCGGTCCTCGAGCTGCTCAAAAAGCTGACAGGACAGAGCGACTACACGTGGGCGGCCGTACTGAAGCTGCTGGACGACCAGGTGCTGCCCCGGGTCAACGCCGACTGGGCTGACCGCCTCACCGCAGCACTCAGGGAGGATCTCTTGAAGACTCTCGGTACGGACGGTGTGCTGCTGTCCGCGAGCGCCCCGCAGCCGGCGCCCTACCACGCCGCGCCGCTGCTGCGACCCTTCAACTTCGCCTACTGGGGGATCTTCAATGTACTCAGGTTCCCCGCTGTGCAGGTACCGTTAGGGCTGAACAGTGAGGGCATTCCCCTCGGCATCCAGGTGGTGGCGGCGCCCGCGCAGGAGGCGCTCTGCCTCGCCGTGGCGGAGCACCTGGCCAGGACCTTCGGGGGCTACGTGCCTCCGTGCAAGATTATAGATACATAG
Protein
MCTTNHGDGRISHSVPGSKTCNIIRGIAFNMIKHAFLVFRTYLDLFIDYIFSLYWEAKRTPLPNLEKKHIILKDDAVTIAAKIRNRQLKSEDLVQMCIERIKIVNPILNAVTDERFEEALEEAREIDRMLDEGLSEEYLKKKPFLGVPFTAKESHAVKGLLHTLGIKARAHVRAHEDAECVRLMRAAGALPLAVTNVPEINKWCETRNMVFGQTNNPYHTGRTTGGSSGGEAALMAAMATPISLCSDIGGSTRMPAFYCGLFGINPTAGYTSLKGSALRSGKDDTMASIGFVSRHTCDLAALTKVILADKAAELDLDRPVDLKTIKYYYLDTAQDLRVSPIQPELREAMDKVISQLSEEASSARRYQHAGLDHMYALWRHAMTREADVFGRLLTNNQGEASAVLELLKKLTGQSDYTWAAVLKLLDDQVLPRVNADWADRLTAALREDLLKTLGTDGVLLSASAPQPAPYHAAPLLRPFNFAYWGIFNVLRFPAVQVPLGLNSEGIPLGIQVVAAPAQEALCLAVAEHLARTFGGYVPPCKIIDT
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
H9JCU8
A0A212EY88
A0A0N0P9X0
A0A194R4Y0
S4PHQ7
A0A2H1W8P6
+ More
A0A2W1BI98 A0A194R5S2 I4DM64 I4DNE7 A0A0N1IMZ1 S4PJ70 A0A212FKU7 A0A224XAH8 H9JD27 A0A023FBJ4 A0A0V0G5H4 A0A0P4VTM9 A0A0C9R1J7 A0A067RTD2 T1HXG2 A0A182GHH5 A0A182GY52 A0A1B6LTD9 A0A182XCG2 A0A182NB16 A0A182ICP2 Q7Q1V4 A0A182UF77 F4WR03 A0A182QBX8 A0A182V4G2 A0A182L615 A0A2J7Q5J7 A0A2R7WWU6 A0A182M016 Q17M12 A0A182VTS9 E2BQI5 E0VE66 A0A182K203 A0A084VNJ4 A0A182Y8E2 A0A182RCC8 A0A154PIB0 A0A182PAS1 A0A151I018 A0A182F2P8 A0A2A3E4A9 A0A195FV19 A0A1Q3G412 A0A151WIF4 A0A2M4BKF0 A0A151IVY7 A0A2M4BK97 A0A026WGB3 A0A1J1HG06 A0A2M4BKC2 A0A2M4AKE3 A0A2M4AK13 R4WD06 A0A2H8TUZ2 W5JC12 A0A088A8Y7 A0A1B6C263 A0A1B6DWW8 A0A182IZD7 K7IUR9 A0A2S2QR36 B4QUX1 A0A0L0BX96 A0A0L7R3H9 D6WXE1 A0A1L8DRM1 B4ICA3 J9JLM2 A0A336KKN6 A0A3Q0IUX8 A0A0A9XIE0 A0A1L8DRC2 A0A0K8TAR1 A0A1Y1KK56 Q9VBQ5 B4PTK0 J3JZG4 A0A232F5Y6 B4NJE6 A0A1I8Q6W8 B3P6I7 A0A1W4VLS3 B4JHK7 A0A3B0KER4 A0A1B0BFD8 A0A0M5J2K8 A0A1A9VNB6 A0A1A9X225 T1PFU2 B4GF55
A0A2W1BI98 A0A194R5S2 I4DM64 I4DNE7 A0A0N1IMZ1 S4PJ70 A0A212FKU7 A0A224XAH8 H9JD27 A0A023FBJ4 A0A0V0G5H4 A0A0P4VTM9 A0A0C9R1J7 A0A067RTD2 T1HXG2 A0A182GHH5 A0A182GY52 A0A1B6LTD9 A0A182XCG2 A0A182NB16 A0A182ICP2 Q7Q1V4 A0A182UF77 F4WR03 A0A182QBX8 A0A182V4G2 A0A182L615 A0A2J7Q5J7 A0A2R7WWU6 A0A182M016 Q17M12 A0A182VTS9 E2BQI5 E0VE66 A0A182K203 A0A084VNJ4 A0A182Y8E2 A0A182RCC8 A0A154PIB0 A0A182PAS1 A0A151I018 A0A182F2P8 A0A2A3E4A9 A0A195FV19 A0A1Q3G412 A0A151WIF4 A0A2M4BKF0 A0A151IVY7 A0A2M4BK97 A0A026WGB3 A0A1J1HG06 A0A2M4BKC2 A0A2M4AKE3 A0A2M4AK13 R4WD06 A0A2H8TUZ2 W5JC12 A0A088A8Y7 A0A1B6C263 A0A1B6DWW8 A0A182IZD7 K7IUR9 A0A2S2QR36 B4QUX1 A0A0L0BX96 A0A0L7R3H9 D6WXE1 A0A1L8DRM1 B4ICA3 J9JLM2 A0A336KKN6 A0A3Q0IUX8 A0A0A9XIE0 A0A1L8DRC2 A0A0K8TAR1 A0A1Y1KK56 Q9VBQ5 B4PTK0 J3JZG4 A0A232F5Y6 B4NJE6 A0A1I8Q6W8 B3P6I7 A0A1W4VLS3 B4JHK7 A0A3B0KER4 A0A1B0BFD8 A0A0M5J2K8 A0A1A9VNB6 A0A1A9X225 T1PFU2 B4GF55
Pubmed
19121390
22118469
26354079
23622113
28756777
22651552
+ More
25474469 27129103 24845553 26483478 12364791 14747013 17210077 21719571 20966253 17510324 20798317 20566863 24438588 25244985 24508170 30249741 23691247 20920257 23761445 20075255 17994087 26108605 18362917 19820115 25401762 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22516182 23537049 28648823
25474469 27129103 24845553 26483478 12364791 14747013 17210077 21719571 20966253 17510324 20798317 20566863 24438588 25244985 24508170 30249741 23691247 20920257 23761445 20075255 17994087 26108605 18362917 19820115 25401762 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22516182 23537049 28648823
EMBL
BABH01027727
AGBW02011586
OWR46450.1
KQ459072
KPJ03855.1
KQ460685
+ More
KPJ12863.1 GAIX01000394 JAA92166.1 ODYU01007041 SOQ49440.1 KZ150177 PZC72556.1 KPJ12864.1 AK402382 BAM19004.1 AK402871 BAM19437.1 KPJ03854.1 GAIX01005115 JAA87445.1 AGBW02008003 OWR54319.1 GFTR01007039 JAW09387.1 BABH01027728 GBBI01000343 JAC18369.1 GECL01003517 JAP02607.1 GDKW01001260 JAI55335.1 GBYB01001835 JAG71602.1 KK852428 KDR24070.1 ACPB03006226 JXUM01063734 JXUM01063735 KQ562265 KXJ76273.1 JXUM01096859 JXUM01096860 KQ564297 KXJ72471.1 GEBQ01013098 JAT26879.1 APCN01005736 AAAB01008980 EAA14577.4 GL888280 EGI63361.1 AXCN02000647 NEVH01017557 PNF23845.1 KK855826 PTY24019.1 AXCM01003770 CH477209 EAT47732.1 GL449769 EFN82043.1 DS235088 EEB11672.1 ATLV01014759 KE524984 KFB39538.1 KQ434924 KZC11579.1 KQ976628 KYM78900.1 KZ288379 PBC26567.1 KQ981261 KYN44137.1 GFDL01000502 JAV34543.1 KQ983089 KYQ47618.1 GGFJ01004312 MBW53453.1 KQ980882 KYN11943.1 GGFJ01004311 MBW53452.1 KK107238 QOIP01000007 EZA54716.1 RLU20207.1 CVRI01000001 CRK86482.1 GGFJ01004313 MBW53454.1 GGFK01007932 MBW41253.1 GGFK01007766 MBW41087.1 AK417298 BAN20513.1 GFXV01005193 MBW16998.1 ADMH02001646 ETN61596.1 GEDC01029691 JAS07607.1 GEDC01007137 JAS30161.1 AAZX01005919 GGMS01010379 MBY79582.1 CM000364 EDX14437.1 JRES01001197 KNC24667.1 KQ414663 KOC65383.1 KQ971361 EFA07993.1 GFDF01004968 JAV09116.1 CH480828 EDW44999.1 ABLF02034972 ABLF02034978 UFQS01000574 UFQT01000574 SSX05049.1 SSX25411.1 GBHO01023860 JAG19744.1 GFDF01005160 JAV08924.1 GBRD01003212 JAG62609.1 GEZM01086074 JAV59796.1 AE014297 AY058674 AAF56474.1 AAL13903.1 CM000160 EDW98740.2 APGK01019841 APGK01053910 BT128646 KB741239 KB740134 KB632275 AEE63603.1 ENN72115.1 ENN81277.1 ERL91089.1 NNAY01000905 OXU25992.1 CH964272 EDW84977.1 CH954182 EDV53657.1 CH916369 EDV92834.1 OUUW01000007 SPP83551.1 JXJN01013383 CP012526 ALC47998.1 KA647687 AFP62316.1 CH479182 EDW34240.1
KPJ12863.1 GAIX01000394 JAA92166.1 ODYU01007041 SOQ49440.1 KZ150177 PZC72556.1 KPJ12864.1 AK402382 BAM19004.1 AK402871 BAM19437.1 KPJ03854.1 GAIX01005115 JAA87445.1 AGBW02008003 OWR54319.1 GFTR01007039 JAW09387.1 BABH01027728 GBBI01000343 JAC18369.1 GECL01003517 JAP02607.1 GDKW01001260 JAI55335.1 GBYB01001835 JAG71602.1 KK852428 KDR24070.1 ACPB03006226 JXUM01063734 JXUM01063735 KQ562265 KXJ76273.1 JXUM01096859 JXUM01096860 KQ564297 KXJ72471.1 GEBQ01013098 JAT26879.1 APCN01005736 AAAB01008980 EAA14577.4 GL888280 EGI63361.1 AXCN02000647 NEVH01017557 PNF23845.1 KK855826 PTY24019.1 AXCM01003770 CH477209 EAT47732.1 GL449769 EFN82043.1 DS235088 EEB11672.1 ATLV01014759 KE524984 KFB39538.1 KQ434924 KZC11579.1 KQ976628 KYM78900.1 KZ288379 PBC26567.1 KQ981261 KYN44137.1 GFDL01000502 JAV34543.1 KQ983089 KYQ47618.1 GGFJ01004312 MBW53453.1 KQ980882 KYN11943.1 GGFJ01004311 MBW53452.1 KK107238 QOIP01000007 EZA54716.1 RLU20207.1 CVRI01000001 CRK86482.1 GGFJ01004313 MBW53454.1 GGFK01007932 MBW41253.1 GGFK01007766 MBW41087.1 AK417298 BAN20513.1 GFXV01005193 MBW16998.1 ADMH02001646 ETN61596.1 GEDC01029691 JAS07607.1 GEDC01007137 JAS30161.1 AAZX01005919 GGMS01010379 MBY79582.1 CM000364 EDX14437.1 JRES01001197 KNC24667.1 KQ414663 KOC65383.1 KQ971361 EFA07993.1 GFDF01004968 JAV09116.1 CH480828 EDW44999.1 ABLF02034972 ABLF02034978 UFQS01000574 UFQT01000574 SSX05049.1 SSX25411.1 GBHO01023860 JAG19744.1 GFDF01005160 JAV08924.1 GBRD01003212 JAG62609.1 GEZM01086074 JAV59796.1 AE014297 AY058674 AAF56474.1 AAL13903.1 CM000160 EDW98740.2 APGK01019841 APGK01053910 BT128646 KB741239 KB740134 KB632275 AEE63603.1 ENN72115.1 ENN81277.1 ERL91089.1 NNAY01000905 OXU25992.1 CH964272 EDW84977.1 CH954182 EDV53657.1 CH916369 EDV92834.1 OUUW01000007 SPP83551.1 JXJN01013383 CP012526 ALC47998.1 KA647687 AFP62316.1 CH479182 EDW34240.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000027135
UP000015103
+ More
UP000069940 UP000249989 UP000076407 UP000075884 UP000075840 UP000007062 UP000075902 UP000007755 UP000075886 UP000075903 UP000075882 UP000235965 UP000075883 UP000008820 UP000075920 UP000008237 UP000009046 UP000075881 UP000030765 UP000076408 UP000075900 UP000076502 UP000075885 UP000078540 UP000069272 UP000242457 UP000078541 UP000075809 UP000078492 UP000053097 UP000279307 UP000183832 UP000000673 UP000005203 UP000075880 UP000002358 UP000000304 UP000037069 UP000053825 UP000007266 UP000001292 UP000007819 UP000079169 UP000000803 UP000002282 UP000019118 UP000030742 UP000215335 UP000007798 UP000095300 UP000008711 UP000192221 UP000001070 UP000268350 UP000092460 UP000092553 UP000078200 UP000091820 UP000008744
UP000069940 UP000249989 UP000076407 UP000075884 UP000075840 UP000007062 UP000075902 UP000007755 UP000075886 UP000075903 UP000075882 UP000235965 UP000075883 UP000008820 UP000075920 UP000008237 UP000009046 UP000075881 UP000030765 UP000076408 UP000075900 UP000076502 UP000075885 UP000078540 UP000069272 UP000242457 UP000078541 UP000075809 UP000078492 UP000053097 UP000279307 UP000183832 UP000000673 UP000005203 UP000075880 UP000002358 UP000000304 UP000037069 UP000053825 UP000007266 UP000001292 UP000007819 UP000079169 UP000000803 UP000002282 UP000019118 UP000030742 UP000215335 UP000007798 UP000095300 UP000008711 UP000192221 UP000001070 UP000268350 UP000092460 UP000092553 UP000078200 UP000091820 UP000008744
Interpro
Gene 3D
ProteinModelPortal
H9JCU8
A0A212EY88
A0A0N0P9X0
A0A194R4Y0
S4PHQ7
A0A2H1W8P6
+ More
A0A2W1BI98 A0A194R5S2 I4DM64 I4DNE7 A0A0N1IMZ1 S4PJ70 A0A212FKU7 A0A224XAH8 H9JD27 A0A023FBJ4 A0A0V0G5H4 A0A0P4VTM9 A0A0C9R1J7 A0A067RTD2 T1HXG2 A0A182GHH5 A0A182GY52 A0A1B6LTD9 A0A182XCG2 A0A182NB16 A0A182ICP2 Q7Q1V4 A0A182UF77 F4WR03 A0A182QBX8 A0A182V4G2 A0A182L615 A0A2J7Q5J7 A0A2R7WWU6 A0A182M016 Q17M12 A0A182VTS9 E2BQI5 E0VE66 A0A182K203 A0A084VNJ4 A0A182Y8E2 A0A182RCC8 A0A154PIB0 A0A182PAS1 A0A151I018 A0A182F2P8 A0A2A3E4A9 A0A195FV19 A0A1Q3G412 A0A151WIF4 A0A2M4BKF0 A0A151IVY7 A0A2M4BK97 A0A026WGB3 A0A1J1HG06 A0A2M4BKC2 A0A2M4AKE3 A0A2M4AK13 R4WD06 A0A2H8TUZ2 W5JC12 A0A088A8Y7 A0A1B6C263 A0A1B6DWW8 A0A182IZD7 K7IUR9 A0A2S2QR36 B4QUX1 A0A0L0BX96 A0A0L7R3H9 D6WXE1 A0A1L8DRM1 B4ICA3 J9JLM2 A0A336KKN6 A0A3Q0IUX8 A0A0A9XIE0 A0A1L8DRC2 A0A0K8TAR1 A0A1Y1KK56 Q9VBQ5 B4PTK0 J3JZG4 A0A232F5Y6 B4NJE6 A0A1I8Q6W8 B3P6I7 A0A1W4VLS3 B4JHK7 A0A3B0KER4 A0A1B0BFD8 A0A0M5J2K8 A0A1A9VNB6 A0A1A9X225 T1PFU2 B4GF55
A0A2W1BI98 A0A194R5S2 I4DM64 I4DNE7 A0A0N1IMZ1 S4PJ70 A0A212FKU7 A0A224XAH8 H9JD27 A0A023FBJ4 A0A0V0G5H4 A0A0P4VTM9 A0A0C9R1J7 A0A067RTD2 T1HXG2 A0A182GHH5 A0A182GY52 A0A1B6LTD9 A0A182XCG2 A0A182NB16 A0A182ICP2 Q7Q1V4 A0A182UF77 F4WR03 A0A182QBX8 A0A182V4G2 A0A182L615 A0A2J7Q5J7 A0A2R7WWU6 A0A182M016 Q17M12 A0A182VTS9 E2BQI5 E0VE66 A0A182K203 A0A084VNJ4 A0A182Y8E2 A0A182RCC8 A0A154PIB0 A0A182PAS1 A0A151I018 A0A182F2P8 A0A2A3E4A9 A0A195FV19 A0A1Q3G412 A0A151WIF4 A0A2M4BKF0 A0A151IVY7 A0A2M4BK97 A0A026WGB3 A0A1J1HG06 A0A2M4BKC2 A0A2M4AKE3 A0A2M4AK13 R4WD06 A0A2H8TUZ2 W5JC12 A0A088A8Y7 A0A1B6C263 A0A1B6DWW8 A0A182IZD7 K7IUR9 A0A2S2QR36 B4QUX1 A0A0L0BX96 A0A0L7R3H9 D6WXE1 A0A1L8DRM1 B4ICA3 J9JLM2 A0A336KKN6 A0A3Q0IUX8 A0A0A9XIE0 A0A1L8DRC2 A0A0K8TAR1 A0A1Y1KK56 Q9VBQ5 B4PTK0 J3JZG4 A0A232F5Y6 B4NJE6 A0A1I8Q6W8 B3P6I7 A0A1W4VLS3 B4JHK7 A0A3B0KER4 A0A1B0BFD8 A0A0M5J2K8 A0A1A9VNB6 A0A1A9X225 T1PFU2 B4GF55
PDB
3A2P
E-value=3.34432e-23,
Score=269
Ontologies
KEGG
GO
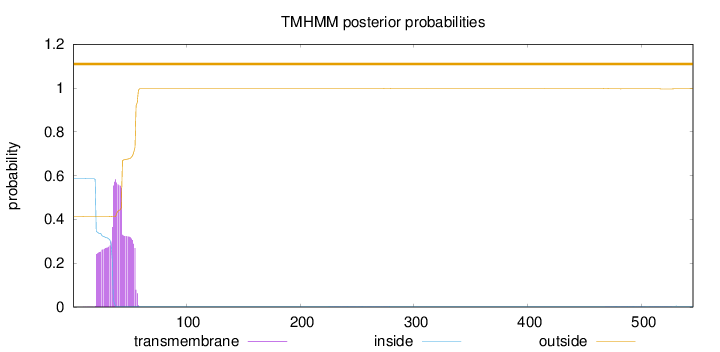
Topology
Length:
545
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.51387
Exp number, first 60 AAs:
12.46052
Total prob of N-in:
0.58615
POSSIBLE N-term signal
sequence
outside
1 - 545
Population Genetic Test Statistics
Pi
250.087867
Theta
171.957922
Tajima's D
1.578482
CLR
0.18447
CSRT
0.806559672016399
Interpretation
Uncertain
