Gene
KWMTBOMO01086
Pre Gene Modal
BGIBMGA007423
Annotation
PREDICTED:_S-adenosylmethionine_decarboxylase_proenzyme_[Bombyx_mori]
Full name
S-adenosylmethionine decarboxylase proenzyme
Location in the cell
Mitochondrial Reliability : 1.038
Sequence
CDS
ATGTTTGTTTCACGAAGACGCTGGATACTGAAGACATGCGGGACGACCACCCCGCTGCGGTGCGTCCGCGCTGTGCTGGCTCTCGCACAGGACGTGGCGGGCCATTCGTGCGTGCACAACGTGTTCTACTCCAGGAGAGAATTCGCCCGTCCCGAAGTCCAGCTCAAACCGCACGACAACTTTGATTCAGAGGTAAAACTTCTGGATTCATTTTTCGGGGATGGGCGCGCCTATATCATGGGCCCGGAAAAAGACTGTTGGTATTTGTATACCCTGCTGCCGTTGGAAGGTACAGTGGCAGCGGTGGAGAAAGAGCAGCAGGAAGCTGAGTCGGGACGCGGGGAGCCCGACCAGACCATTGAGATTCTCATGTCCGATCTCGATCCTCAGGTCATGGACATATTTACCCGGGAACATTCCCACGATGCTGCGTATGCCACTAAAGAGTCTGGCATAGACAAAATATTACCAGGAATGGTCATAGACGATTTCCTGTTTGACCCCTGCGGCTACTCTATGAACGGGGTCGCTAAAGATGGTTGCTACATGACGATCCACATAACGCCCGAGAGGGCCTGCTCGTACGTGTCGTTCGAGTCCAACGTGCCCGTGTCCAGCTACGACGAGGTGATAGCGCGCGTGCTGGCCGCCTTCCGCCCCGCCAAGTTCGTGCTCACCATCTTCGCCACGCCGGACTCCCCGGCGGCGGACGCGTACCGGCAGCTGCGCAAGTACCGCTCGCTGAGCGCGTACGAGGAGCAGGAGGCGCAGTACTGCCGGTTCTCTGGGTACGAGCTGCAGTACGCGCTGTACTGCAATATATGGCGAGCTCGGCGTGTAGCGCGCAGACTCGCGGGCCGGGGCGGGTCGCCGGCGCAGGCCGCTCCTGAGCGCGGGCCGAAATTATTATCGATGATTTTAAAAATCAAATAA
Protein
MFVSRRRWILKTCGTTTPLRCVRAVLALAQDVAGHSCVHNVFYSRREFARPEVQLKPHDNFDSEVKLLDSFFGDGRAYIMGPEKDCWYLYTLLPLEGTVAAVEKEQQEAESGRGEPDQTIEILMSDLDPQVMDIFTREHSHDAAYATKESGIDKILPGMVIDDFLFDPCGYSMNGVAKDGCYMTIHITPERACSYVSFESNVPVSSYDEVIARVLAAFRPAKFVLTIFATPDSPAADAYRQLRKYRSLSAYEEQEAQYCRFSGYELQYALYCNIWRARRVARRLAGRGGSPAQAAPERGPKLLSMILKIK
Summary
Catalytic Activity
H(+) + S-adenosyl-L-methionine = CO2 + S-adenosyl 3-(methylsulfanyl)propylamine
Cofactor
pyruvate
Similarity
Belongs to the eukaryotic AdoMetDC family.
Keywords
Autocatalytic cleavage
Complete proteome
Decarboxylase
Lyase
Polyamine biosynthesis
Pyruvate
Reference proteome
S-adenosyl-L-methionine
Schiff base
Spermidine biosynthesis
Zymogen
Feature
chain S-adenosylmethionine decarboxylase beta chain
Uniprot
H9JD26
A0A2A4JC49
A0A1E1W7Y2
A0A0N0PA84
A0A194R5A8
A0A2H1WKU2
+ More
A0A212EY80 A0A2J7Q5L0 A0A067RM14 A0A2J7Q5M2 A0A1B0CJG1 A0A1L8DM26 A0A1J1HGF6 A0A1J1HF75 U5EYZ7 Q7Q1V3 A0A182VTT1 A0A182MF58 A0A084VNJ2 T1E1J8 E0VW29 D6WXD9 A0A1Y1MAF3 E2AXS3 A0A088AM45 A0A310SGW2 A0A0C9PI02 A0A1A9W3Z7 A0A023F2T2 A0A336LQ82 E9J4Y0 V9IDP3 A0A1B6L689 F4WFI4 A0A3L8DI51 A0A151IR57 A0A195CWX1 A0A158NS80 A0A151WPQ1 A0A1B6HA52 A0A1B6EP53 A0A1B6CYT8 A0A026WHS0 A0A195BRW1 E2B488 A0A151JVI6 A0A336LU59 A0A1A9XE11 A0A1B0BVY5 A0A0A9WGW1 T1DI65 A0A1B6D7X2 A0A1B0AC17 D3TN20 A0A0L0C121 B4M7A4 A0A1A9ULE1 T1GVF3 A0A0M4ER25 T1IQC5 B4JP06 A0A0B4RYL6 B4HWL2 A0A1I8N2B1 B4Q8U4 A0A1I8Q5N1 Q58CK1 N6T6S3 T1PEA9 P91931 T1HB17 B3N9L8 A0A0M8ZRC1 A0A0J9R0X2 A0A0T6B3E6 A0A1V9XI98 A0A0A1XDF2 A0A0K2TJT2 B4L5R4 A0A0K8WKU1 A0A131XZM8 A0A0K8VQN0 B4G7J1 B4NZA3 N6U2Y9 A0A2A3E767 A0A034W9U5 T1KY54 A0A154PC46 T1KXT0 A0A0M4EKE2 W8CDS7 A0A0K8RQT7 A0A0P4WRQ4 A0A2R5LGK5 A0A2H8TDI5 A0A3B0K197 A0A293N3G4
A0A212EY80 A0A2J7Q5L0 A0A067RM14 A0A2J7Q5M2 A0A1B0CJG1 A0A1L8DM26 A0A1J1HGF6 A0A1J1HF75 U5EYZ7 Q7Q1V3 A0A182VTT1 A0A182MF58 A0A084VNJ2 T1E1J8 E0VW29 D6WXD9 A0A1Y1MAF3 E2AXS3 A0A088AM45 A0A310SGW2 A0A0C9PI02 A0A1A9W3Z7 A0A023F2T2 A0A336LQ82 E9J4Y0 V9IDP3 A0A1B6L689 F4WFI4 A0A3L8DI51 A0A151IR57 A0A195CWX1 A0A158NS80 A0A151WPQ1 A0A1B6HA52 A0A1B6EP53 A0A1B6CYT8 A0A026WHS0 A0A195BRW1 E2B488 A0A151JVI6 A0A336LU59 A0A1A9XE11 A0A1B0BVY5 A0A0A9WGW1 T1DI65 A0A1B6D7X2 A0A1B0AC17 D3TN20 A0A0L0C121 B4M7A4 A0A1A9ULE1 T1GVF3 A0A0M4ER25 T1IQC5 B4JP06 A0A0B4RYL6 B4HWL2 A0A1I8N2B1 B4Q8U4 A0A1I8Q5N1 Q58CK1 N6T6S3 T1PEA9 P91931 T1HB17 B3N9L8 A0A0M8ZRC1 A0A0J9R0X2 A0A0T6B3E6 A0A1V9XI98 A0A0A1XDF2 A0A0K2TJT2 B4L5R4 A0A0K8WKU1 A0A131XZM8 A0A0K8VQN0 B4G7J1 B4NZA3 N6U2Y9 A0A2A3E767 A0A034W9U5 T1KY54 A0A154PC46 T1KXT0 A0A0M4EKE2 W8CDS7 A0A0K8RQT7 A0A0P4WRQ4 A0A2R5LGK5 A0A2H8TDI5 A0A3B0K197 A0A293N3G4
EC Number
4.1.1.50
Pubmed
19121390
26354079
22118469
24845553
12364791
24438588
+ More
24330624 20566863 18362917 19820115 28004739 20798317 25474469 21282665 21719571 30249741 21347285 24508170 25401762 20353571 26108605 17994087 25592131 25315136 23537049 9435790 10731132 12537572 22936249 28327890 25830018 17550304 25348373 24495485
24330624 20566863 18362917 19820115 28004739 20798317 25474469 21282665 21719571 30249741 21347285 24508170 25401762 20353571 26108605 17994087 25592131 25315136 23537049 9435790 10731132 12537572 22936249 28327890 25830018 17550304 25348373 24495485
EMBL
BABH01027726
NWSH01002172
PCG68973.1
GDQN01007942
JAT83112.1
KQ459072
+ More
KPJ03856.1 KQ460685 KPJ12862.1 ODYU01009000 SOQ53074.1 AGBW02011585 OWR46452.1 NEVH01017557 PNF23869.1 KK852428 KDR24068.1 PNF23870.1 AJWK01014578 AJWK01014579 AJWK01014580 GFDF01006623 JAV07461.1 CVRI01000001 CRK86484.1 CRK86483.1 GANO01001701 JAB58170.1 AAAB01008980 EAA14565.4 AXCM01003770 ATLV01014759 KE524984 KFB39536.1 GALA01001599 JAA93253.1 DS235816 EEB17585.1 KQ971361 EFA07994.1 GEZM01036297 JAV82819.1 GL443653 EFN61772.1 KQ761402 OAD57564.1 GBYB01000518 GBYB01000520 JAG70285.1 JAG70287.1 GBBI01003072 JAC15640.1 UFQT01000114 SSX20224.1 GL768121 EFZ12126.1 JR038906 AEY58449.1 GEBQ01020739 JAT19238.1 GL888116 EGI67056.1 QOIP01000007 RLU20124.1 KQ981147 KYN09106.1 KQ977171 KYN05166.1 ADTU01024665 ADTU01024666 ADTU01024667 ADTU01024668 KQ982883 KYQ49665.1 GECU01036138 GECU01017690 JAS71568.1 JAS90016.1 GECZ01030062 GECZ01014615 JAS39707.1 JAS55154.1 GEDC01018681 JAS18617.1 KK107238 EZA54604.1 KQ976421 KYM89102.1 GL445515 EFN89505.1 KQ981688 KYN37915.1 SSX20223.1 JXJN01021533 GBHO01036570 GBRD01009652 JAG07034.1 JAG56172.1 GALA01001116 JAA93736.1 GEDC01015526 JAS21772.1 CCAG010016028 EZ422822 ADD19098.1 JRES01001143 KNC25159.1 CH940653 EDW62671.2 CAQQ02122322 CP012528 ALC48502.1 JH431303 CH916371 EDV92449.1 KJ136120 AIZ03433.1 CH480818 EDW52407.1 CM000361 EDX04509.1 BT021946 AAX51651.1 APGK01041465 KB740994 ENN75934.1 KA646283 AFP60912.1 Y11216 Y11820 AE014134 ACPB03011086 ACPB03011087 ACPB03011088 ACPB03011089 CH954177 EDV58513.2 KQ435955 KOX68063.1 CM002910 KMY89494.1 LJIG01015993 KRT81936.1 MNPL01010499 OQR73093.1 GBXI01005794 JAD08498.1 HACA01008823 CDW26184.1 CH933811 EDW06523.1 GDHF01000602 JAI51712.1 GEFM01004725 JAP71071.1 GDHF01027931 GDHF01011161 JAI24383.1 JAI41153.1 CH479180 EDW28391.1 CM000157 EDW88798.1 KB630631 KB632280 ENN75935.1 ERL83742.1 ERL91300.1 KZ288347 PBC27555.1 GAKP01007474 GAKP01007473 JAC51479.1 CAEY01000699 KQ434870 KZC09476.1 CAEY01000697 CP012526 ALC47646.1 GAMC01001426 JAC05130.1 GADI01000824 JAA72984.1 GDRN01040355 JAI67159.1 GGLE01004530 MBY08656.1 GFXV01000371 MBW12176.1 OUUW01000006 SPP81630.1 GFWV01021197 MAA45925.1
KPJ03856.1 KQ460685 KPJ12862.1 ODYU01009000 SOQ53074.1 AGBW02011585 OWR46452.1 NEVH01017557 PNF23869.1 KK852428 KDR24068.1 PNF23870.1 AJWK01014578 AJWK01014579 AJWK01014580 GFDF01006623 JAV07461.1 CVRI01000001 CRK86484.1 CRK86483.1 GANO01001701 JAB58170.1 AAAB01008980 EAA14565.4 AXCM01003770 ATLV01014759 KE524984 KFB39536.1 GALA01001599 JAA93253.1 DS235816 EEB17585.1 KQ971361 EFA07994.1 GEZM01036297 JAV82819.1 GL443653 EFN61772.1 KQ761402 OAD57564.1 GBYB01000518 GBYB01000520 JAG70285.1 JAG70287.1 GBBI01003072 JAC15640.1 UFQT01000114 SSX20224.1 GL768121 EFZ12126.1 JR038906 AEY58449.1 GEBQ01020739 JAT19238.1 GL888116 EGI67056.1 QOIP01000007 RLU20124.1 KQ981147 KYN09106.1 KQ977171 KYN05166.1 ADTU01024665 ADTU01024666 ADTU01024667 ADTU01024668 KQ982883 KYQ49665.1 GECU01036138 GECU01017690 JAS71568.1 JAS90016.1 GECZ01030062 GECZ01014615 JAS39707.1 JAS55154.1 GEDC01018681 JAS18617.1 KK107238 EZA54604.1 KQ976421 KYM89102.1 GL445515 EFN89505.1 KQ981688 KYN37915.1 SSX20223.1 JXJN01021533 GBHO01036570 GBRD01009652 JAG07034.1 JAG56172.1 GALA01001116 JAA93736.1 GEDC01015526 JAS21772.1 CCAG010016028 EZ422822 ADD19098.1 JRES01001143 KNC25159.1 CH940653 EDW62671.2 CAQQ02122322 CP012528 ALC48502.1 JH431303 CH916371 EDV92449.1 KJ136120 AIZ03433.1 CH480818 EDW52407.1 CM000361 EDX04509.1 BT021946 AAX51651.1 APGK01041465 KB740994 ENN75934.1 KA646283 AFP60912.1 Y11216 Y11820 AE014134 ACPB03011086 ACPB03011087 ACPB03011088 ACPB03011089 CH954177 EDV58513.2 KQ435955 KOX68063.1 CM002910 KMY89494.1 LJIG01015993 KRT81936.1 MNPL01010499 OQR73093.1 GBXI01005794 JAD08498.1 HACA01008823 CDW26184.1 CH933811 EDW06523.1 GDHF01000602 JAI51712.1 GEFM01004725 JAP71071.1 GDHF01027931 GDHF01011161 JAI24383.1 JAI41153.1 CH479180 EDW28391.1 CM000157 EDW88798.1 KB630631 KB632280 ENN75935.1 ERL83742.1 ERL91300.1 KZ288347 PBC27555.1 GAKP01007474 GAKP01007473 JAC51479.1 CAEY01000699 KQ434870 KZC09476.1 CAEY01000697 CP012526 ALC47646.1 GAMC01001426 JAC05130.1 GADI01000824 JAA72984.1 GDRN01040355 JAI67159.1 GGLE01004530 MBY08656.1 GFXV01000371 MBW12176.1 OUUW01000006 SPP81630.1 GFWV01021197 MAA45925.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000235965
+ More
UP000027135 UP000092461 UP000183832 UP000007062 UP000075920 UP000075883 UP000030765 UP000009046 UP000007266 UP000000311 UP000005203 UP000091820 UP000007755 UP000279307 UP000078492 UP000078542 UP000005205 UP000075809 UP000053097 UP000078540 UP000008237 UP000078541 UP000092443 UP000092460 UP000092445 UP000092444 UP000037069 UP000008792 UP000078200 UP000015102 UP000092553 UP000001070 UP000001292 UP000095301 UP000000304 UP000095300 UP000019118 UP000000803 UP000015103 UP000008711 UP000053105 UP000192247 UP000009192 UP000008744 UP000002282 UP000030742 UP000242457 UP000015104 UP000076502 UP000268350
UP000027135 UP000092461 UP000183832 UP000007062 UP000075920 UP000075883 UP000030765 UP000009046 UP000007266 UP000000311 UP000005203 UP000091820 UP000007755 UP000279307 UP000078492 UP000078542 UP000005205 UP000075809 UP000053097 UP000078540 UP000008237 UP000078541 UP000092443 UP000092460 UP000092445 UP000092444 UP000037069 UP000008792 UP000078200 UP000015102 UP000092553 UP000001070 UP000001292 UP000095301 UP000000304 UP000095300 UP000019118 UP000000803 UP000015103 UP000008711 UP000053105 UP000192247 UP000009192 UP000008744 UP000002282 UP000030742 UP000242457 UP000015104 UP000076502 UP000268350
Pfam
PF01536 SAM_decarbox
Interpro
SUPFAM
SSF56276
SSF56276
ProteinModelPortal
H9JD26
A0A2A4JC49
A0A1E1W7Y2
A0A0N0PA84
A0A194R5A8
A0A2H1WKU2
+ More
A0A212EY80 A0A2J7Q5L0 A0A067RM14 A0A2J7Q5M2 A0A1B0CJG1 A0A1L8DM26 A0A1J1HGF6 A0A1J1HF75 U5EYZ7 Q7Q1V3 A0A182VTT1 A0A182MF58 A0A084VNJ2 T1E1J8 E0VW29 D6WXD9 A0A1Y1MAF3 E2AXS3 A0A088AM45 A0A310SGW2 A0A0C9PI02 A0A1A9W3Z7 A0A023F2T2 A0A336LQ82 E9J4Y0 V9IDP3 A0A1B6L689 F4WFI4 A0A3L8DI51 A0A151IR57 A0A195CWX1 A0A158NS80 A0A151WPQ1 A0A1B6HA52 A0A1B6EP53 A0A1B6CYT8 A0A026WHS0 A0A195BRW1 E2B488 A0A151JVI6 A0A336LU59 A0A1A9XE11 A0A1B0BVY5 A0A0A9WGW1 T1DI65 A0A1B6D7X2 A0A1B0AC17 D3TN20 A0A0L0C121 B4M7A4 A0A1A9ULE1 T1GVF3 A0A0M4ER25 T1IQC5 B4JP06 A0A0B4RYL6 B4HWL2 A0A1I8N2B1 B4Q8U4 A0A1I8Q5N1 Q58CK1 N6T6S3 T1PEA9 P91931 T1HB17 B3N9L8 A0A0M8ZRC1 A0A0J9R0X2 A0A0T6B3E6 A0A1V9XI98 A0A0A1XDF2 A0A0K2TJT2 B4L5R4 A0A0K8WKU1 A0A131XZM8 A0A0K8VQN0 B4G7J1 B4NZA3 N6U2Y9 A0A2A3E767 A0A034W9U5 T1KY54 A0A154PC46 T1KXT0 A0A0M4EKE2 W8CDS7 A0A0K8RQT7 A0A0P4WRQ4 A0A2R5LGK5 A0A2H8TDI5 A0A3B0K197 A0A293N3G4
A0A212EY80 A0A2J7Q5L0 A0A067RM14 A0A2J7Q5M2 A0A1B0CJG1 A0A1L8DM26 A0A1J1HGF6 A0A1J1HF75 U5EYZ7 Q7Q1V3 A0A182VTT1 A0A182MF58 A0A084VNJ2 T1E1J8 E0VW29 D6WXD9 A0A1Y1MAF3 E2AXS3 A0A088AM45 A0A310SGW2 A0A0C9PI02 A0A1A9W3Z7 A0A023F2T2 A0A336LQ82 E9J4Y0 V9IDP3 A0A1B6L689 F4WFI4 A0A3L8DI51 A0A151IR57 A0A195CWX1 A0A158NS80 A0A151WPQ1 A0A1B6HA52 A0A1B6EP53 A0A1B6CYT8 A0A026WHS0 A0A195BRW1 E2B488 A0A151JVI6 A0A336LU59 A0A1A9XE11 A0A1B0BVY5 A0A0A9WGW1 T1DI65 A0A1B6D7X2 A0A1B0AC17 D3TN20 A0A0L0C121 B4M7A4 A0A1A9ULE1 T1GVF3 A0A0M4ER25 T1IQC5 B4JP06 A0A0B4RYL6 B4HWL2 A0A1I8N2B1 B4Q8U4 A0A1I8Q5N1 Q58CK1 N6T6S3 T1PEA9 P91931 T1HB17 B3N9L8 A0A0M8ZRC1 A0A0J9R0X2 A0A0T6B3E6 A0A1V9XI98 A0A0A1XDF2 A0A0K2TJT2 B4L5R4 A0A0K8WKU1 A0A131XZM8 A0A0K8VQN0 B4G7J1 B4NZA3 N6U2Y9 A0A2A3E767 A0A034W9U5 T1KY54 A0A154PC46 T1KXT0 A0A0M4EKE2 W8CDS7 A0A0K8RQT7 A0A0P4WRQ4 A0A2R5LGK5 A0A2H8TDI5 A0A3B0K197 A0A293N3G4
PDB
1JEN
E-value=8.65576e-57,
Score=556
Ontologies
PATHWAY
GO
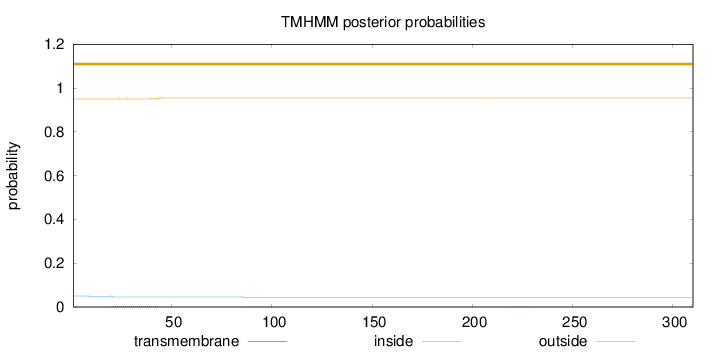
Topology
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11128
Exp number, first 60 AAs:
0.10444
Total prob of N-in:
0.04970
outside
1 - 310
Population Genetic Test Statistics
Pi
227.523773
Theta
147.31258
Tajima's D
1.650903
CLR
0.636109
CSRT
0.812509374531273
Interpretation
Uncertain
