Pre Gene Modal
BGIBMGA007346
Annotation
PREDICTED:_activating_signal_cointegrator_1_complex_subunit_2_isoform_X1_[Amyelois_transitella]
Full name
Activating signal cointegrator 1 complex subunit 2
Alternative Name
ASC-1 complex subunit p100
Trip4 complex subunit p100
Trip4 complex subunit p100
Location in the cell
Nuclear Reliability : 2.019
Sequence
CDS
ATGTCAGATAAACACGTCGAATTTCGTAATCCCGACTACTTGCCAATTGAAGCGGTGTTTTTAACTGTGACGAACGCCGGCGTTATACAACAAATTAAAGCTTTGGATCCGTACTGGGTGGATAATAAGACATTGTCTGTTTATGAAAGACCACCATCAGGCAGCAGTATGGTGATTGGTGCTGTGGACATGTGGAAGACCAAGATGGCAGTATACTTAACAGATTTGAAATGGCTACTGAGTTTGGAATACCATAAGTTCTGGAGTACAGTTCTCTATCACCCGACATGTATGGATACCCTCGTGTCGTTTTTACAAGAAGCCACCCCTCCATATTTACCCCCACACGAAAGTAAAGAGATACAGAATTTATATGAGGAGATTAGAAGATTTGTACTTATAGTTTTTAGTAGACTTGTCACGAATAAAGAGAGCAAATCTTGCTGGATGTCAAAAGAATATATGGGTGACGTACTGTACAATAATTTCATATTCACGATACCGATAATATGGGACATATGTTTGACATATGGTGTGGACAACACAAGGCATGTGTCACGAGTGTTGGACAGTCTCTTCACATTGCAGCCGAGGTATGAGACAGATGCTGTGGCTGCTGTGGCCTTCGTTCATGAGGCTTTTAAATTCATTATCCTACAAGTCAACAAAGATTATGATAGTGATGAGCCACCGAACTTACCTGAGACCTTCAAAGGGTTCAGCGAGATAAGGCGGCCCACAAACAGTAAGACAAAGGATAAACTCACTTTTGAAGTGCTCAAAGATTTAATAATACATTTGTTGGACACCGCATTGACTCTGAGGATATTTATACAAGTGTACCCTAAGGGAGTTAATATATTTAGGAAGACTAATTTTGTTTTAAGTATAGTTCAACTTTACGAATATGGTGTACCGCAGCTTTACGAGAAGCTGGAGGAGGTGGGGGATCAGACCAGCGTCGCGTATACGGAAATAGAGTCGTACCTCGACACGGCTAGAGCTGAACTCATCGATGTATTTAGAGAGATATTGGCGGTTTATAAAAACGCTATATTTAGTGAAGAAGGTAGTAAAGAGAGCCACGTGGAAGATTACCTGGCGGTGATGATGGAAGGTCTCTCCGAAAGGCTTCTCATCAAAGACTACCACAGCTGTTACCCAGTGCACGAGGACCTGGACTTACTGAGGCAGGCGTGTCCTGAAATAGACACCGTGAAAACGGACTTTATATTGCAAGCTGTCTACTCGAGCCTGGACGAGCCAGCGCCGGAGCCCGCCCCGCCCCCCGCCCCCAACACCGCCACCCCCCACATCGGCACGCCCCACCCCGCCCCCTCAGACAACGAGATCCCGGACACCGTCAAGGAGCAGTCGCTGATCAGCGAGGTCCGGGACATCCTGCCGCACCTCGGGGACGGCTTCATACTGAAGTGCTTACAACATTACGGCTTCAATTCCGAACGGGTGATAAACAGCGTGTTGGAAGATAATCTGGCGGAACCCTTGAGAGCACTAGACCGGAGCCTGCCTATAATACCGGAGGACCCGGTCGACAGGCGCTACCTGGAGACCGGCATCCAACGGCTGAACGTGTTTGACGGAGACGAGTTCGACCTCATGTCCCGGGACGACGTGGTCACGGCCCGGATGCACGTCGGCAAGCGGAGGTCCAAGTACAAGGACCTCACGCAAATGCTCGACGACAAGACGGAGCTCCGGGGGCTCGCGGACATCTACGCCAAGTACAGCGTGGTGTGCGACGAGGAGGACATGTACTCCGACGAGTACGACGACACGGACGACTGGCAGCCGCCCGCCGCCGGCGACCACCCGCCCTCCGAGGAATCTGAAGAGGAGCCCGAGCCGGAGGAGGAGAAACCTTCGAGCAGCAGGAACAGGATGGACTTCTGCGTGAACCCCGAGGAGCTCCGGGCGAGGCGAGAGGCCAGCTTCCAGGCCAGGAGGGGCAGAGGATACAGACCTCCTGCAAAGAATGTGGACGTCGTGGGGAAAGCTAAGGGTCAAGGTCAAGAAAAGAACGTCCTCGTGAACAGGGAGAGGAAGGAGAAGAACAAGTCCTCCCGCGGCAACCACAGCAGGAGGCAGGGGGCGCAGTGGAAGCGCACCCAAGGACTTGTGCCGTCGTAA
Protein
MSDKHVEFRNPDYLPIEAVFLTVTNAGVIQQIKALDPYWVDNKTLSVYERPPSGSSMVIGAVDMWKTKMAVYLTDLKWLLSLEYHKFWSTVLYHPTCMDTLVSFLQEATPPYLPPHESKEIQNLYEEIRRFVLIVFSRLVTNKESKSCWMSKEYMGDVLYNNFIFTIPIIWDICLTYGVDNTRHVSRVLDSLFTLQPRYETDAVAAVAFVHEAFKFIILQVNKDYDSDEPPNLPETFKGFSEIRRPTNSKTKDKLTFEVLKDLIIHLLDTALTLRIFIQVYPKGVNIFRKTNFVLSIVQLYEYGVPQLYEKLEEVGDQTSVAYTEIESYLDTARAELIDVFREILAVYKNAIFSEEGSKESHVEDYLAVMMEGLSERLLIKDYHSCYPVHEDLDLLRQACPEIDTVKTDFILQAVYSSLDEPAPEPAPPPAPNTATPHIGTPHPAPSDNEIPDTVKEQSLISEVRDILPHLGDGFILKCLQHYGFNSERVINSVLEDNLAEPLRALDRSLPIIPEDPVDRRYLETGIQRLNVFDGDEFDLMSRDDVVTARMHVGKRRSKYKDLTQMLDDKTELRGLADIYAKYSVVCDEEDMYSDEYDDTDDWQPPAAGDHPPSEESEEEPEPEEEKPSSSRNRMDFCVNPEELRARREASFQARRGRGYRPPAKNVDVVGKAKGQGQEKNVLVNRERKEKNKSSRGNHSRRQGAQWKRTQGLVPS
Summary
Description
Plays a role in DNA damage repair as component of the ASCC complex. Recruits ASCC3 and ALKBH3 to sites of DNA damage by binding to polyubiquitinated proteins that have 'Lys-63'-linked polyubiquitin chains. Part of the ASC-1 complex that enhances NF-kappa-B, SRF and AP1 transactivation.
Subunit
Identified in the ASCC complex that contains ASCC1, ASCC2 and ASCC3. Interacts directly with ASCC3. The ASCC complex interacts with ALKBH3. Interacts (via CUE domain) with 'Lys-63'-linked polyubiquitin chains, but not with 'Lys-48'-linked polyubiquitin chains. Part of the ASC-1 complex, that contains TRIP4, ASCC1, ASCC2 and ASCC3. Interacts with CSRP1. Interacts with PRPF8, a component of the spliceosome.
Similarity
Belongs to the ASCC2 family.
Keywords
Alternative splicing
Complete proteome
DNA damage
DNA repair
Nucleus
Phosphoprotein
Reference proteome
Transcription
Transcription regulation
Feature
chain Activating signal cointegrator 1 complex subunit 2
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JCU9
A0A212F0K6
A0A2A4JAH9
A0A194R5R7
A0A0N0P9X2
A0A0C9QW05
+ More
A0A067R101 A0A195F513 A0A232EE01 K7JAM1 E9IAW0 A0A195BVZ7 A0A158NRK5 A0A195EK54 A0A2J7PDC7 A0A2J7PDE4 A0A0C9QVZ9 F4X1L0 A0A151I8S3 A0A0L7R8S9 A0A0C9PU20 A0A0V0G8Y4 A0A182JD78 A0A1B6LZ56 A0A069DWR0 A0A224X864 A0A034WNB7 Q16FQ3 A0A034WQ05 A0A1B6JBW3 W8C9T6 A0A3L8DQW0 A0A182H8Y8 A0A0M9A8Y7 A0A023EVX1 A0A336KQ35 A0A1B6G460 A0A023EY09 Q7QFD7 A0A1B6E5A1 A0A1B0GQR2 A0A182IAT0 A0A088AR41 A0A182UVX2 A0A1Q3FCG2 A0A182XDS2 A0A2M3Z5Q5 A0A2M3Z5N1 A0A1Q3FE69 A0A084VNV7 A0A1Q3FDB6 A0A2A3EDL8 A0A2M4BFN4 B0XFK9 A0A182U305 A0A0P4VUW3 A0A1S3WJ36 A0A182PBK8 A0A1I8NME4 A0A096LYT5 A0A2U3VX67 A0A0A1X4E1 Q7ZXU8 S7PH01 A0A2Y9I5L4 A0A286ZQL9 A0A182GIZ2 G1PWT1 F6U4G2 W4YDX4 A0A061INR1 E2RS73 G3HP16 M3YP83 K9J2P7 Q91WR3 A0A2Y9L4D5 A0A1L8DHC1 A0A061IMU8 G3TQL6 Q91WR3-3 Q5E9R9 F7B2Z2 M3W1V2 G3VLN4 A0A2U3Y7Z7 A0A3Q0CNE6 A0A2Y9I5D8 A0A2K6BI62 A0A061IQW0 G1U1Z1 Q3UG63 A0A091FXE8 A0A2K5F1L4 A0A3L8S7C0 A0A2Y9RD40
A0A067R101 A0A195F513 A0A232EE01 K7JAM1 E9IAW0 A0A195BVZ7 A0A158NRK5 A0A195EK54 A0A2J7PDC7 A0A2J7PDE4 A0A0C9QVZ9 F4X1L0 A0A151I8S3 A0A0L7R8S9 A0A0C9PU20 A0A0V0G8Y4 A0A182JD78 A0A1B6LZ56 A0A069DWR0 A0A224X864 A0A034WNB7 Q16FQ3 A0A034WQ05 A0A1B6JBW3 W8C9T6 A0A3L8DQW0 A0A182H8Y8 A0A0M9A8Y7 A0A023EVX1 A0A336KQ35 A0A1B6G460 A0A023EY09 Q7QFD7 A0A1B6E5A1 A0A1B0GQR2 A0A182IAT0 A0A088AR41 A0A182UVX2 A0A1Q3FCG2 A0A182XDS2 A0A2M3Z5Q5 A0A2M3Z5N1 A0A1Q3FE69 A0A084VNV7 A0A1Q3FDB6 A0A2A3EDL8 A0A2M4BFN4 B0XFK9 A0A182U305 A0A0P4VUW3 A0A1S3WJ36 A0A182PBK8 A0A1I8NME4 A0A096LYT5 A0A2U3VX67 A0A0A1X4E1 Q7ZXU8 S7PH01 A0A2Y9I5L4 A0A286ZQL9 A0A182GIZ2 G1PWT1 F6U4G2 W4YDX4 A0A061INR1 E2RS73 G3HP16 M3YP83 K9J2P7 Q91WR3 A0A2Y9L4D5 A0A1L8DHC1 A0A061IMU8 G3TQL6 Q91WR3-3 Q5E9R9 F7B2Z2 M3W1V2 G3VLN4 A0A2U3Y7Z7 A0A3Q0CNE6 A0A2Y9I5D8 A0A2K6BI62 A0A061IQW0 G1U1Z1 Q3UG63 A0A091FXE8 A0A2K5F1L4 A0A3L8S7C0 A0A2Y9RD40
Pubmed
19121390
22118469
26354079
24845553
28648823
20075255
+ More
21282665 21347285 21719571 26334808 25348373 17510324 24495485 30249741 26483478 24945155 25474469 12364791 14747013 17210077 24438588 27129103 25830018 27762356 30723633 21993624 19892987 23929341 16341006 21804562 16141072 19468303 15489334 21183079 11282978 16305752 17495919 17975172 21709235 26319212 10349636 11042159 11076861 11217851 12466851 16141073 30282656
21282665 21347285 21719571 26334808 25348373 17510324 24495485 30249741 26483478 24945155 25474469 12364791 14747013 17210077 24438588 27129103 25830018 27762356 30723633 21993624 19892987 23929341 16341006 21804562 16141072 19468303 15489334 21183079 11282978 16305752 17495919 17975172 21709235 26319212 10349636 11042159 11076861 11217851 12466851 16141073 30282656
EMBL
BABH01027721
BABH01027722
AGBW02011066
OWR47279.1
NWSH01002172
PCG68971.1
+ More
KQ460685 KPJ12859.1 KQ459072 KPJ03858.1 GBYB01004832 JAG74599.1 KK853016 KDR12475.1 KQ981805 KYN35548.1 NNAY01005719 OXU16575.1 AAZX01012632 AAZX01017245 GL762073 EFZ22294.1 KQ976403 KYM92136.1 ADTU01002059 KQ978801 KYN28297.1 NEVH01026389 PNF14346.1 PNF14343.1 GBYB01004827 JAG74594.1 GL888531 EGI59668.1 KQ978331 KYM95064.1 KQ414631 KOC67275.1 GBYB01004828 JAG74595.1 GECL01001791 JAP04333.1 GEBQ01011011 JAT28966.1 GBGD01000618 JAC88271.1 GFTR01007859 JAW08567.1 GAKP01003115 JAC55837.1 CH478401 EAT33062.1 GAKP01003114 JAC55838.1 GECU01011193 JAS96513.1 GAMC01006366 JAC00190.1 QOIP01000005 RLU22676.1 JXUM01029787 JXUM01029788 KQ560864 KXJ80582.1 KQ435711 KOX79647.1 GAPW01000357 JAC13241.1 UFQS01000627 UFQT01000627 SSX05525.1 SSX25884.1 GECZ01012666 JAS57103.1 GBBI01004796 JAC13916.1 AAAB01008846 EAA06509.5 GEDC01004195 JAS33103.1 AJVK01017844 AJVK01017845 APCN01001316 GFDL01009836 JAV25209.1 GGFM01003083 MBW23834.1 GGFM01003078 MBW23829.1 GFDL01009263 JAV25782.1 ATLV01014931 KE524996 KFB39651.1 GFDL01009553 JAV25492.1 KZ288289 PBC29266.1 GGFJ01002703 MBW51844.1 DS232933 EDS26872.1 GDKW01000077 JAI56518.1 AYCK01002591 AYCK01002592 AYCK01002593 GBXI01008103 JAD06189.1 BC044110 BC108785 CM004467 AAH44110.1 AAI08786.1 OCT98127.1 KE162863 EPQ09913.1 AEMK02000096 DQIR01199597 HDB55074.1 JXUM01012471 KQ560355 KXJ82864.1 AAPE02038420 AAGJ04021643 KE663652 ERE91092.1 AAEX03014792 JH000563 EGW12303.1 ERE91090.1 AEYP01066060 GABZ01004591 JAA48934.1 AK048876 AK051387 AL606521 BC013537 BC024840 GFDF01008307 JAV05777.1 ERE91091.1 BT020851 AAX08868.1 AANG04000189 AEFK01073279 AEFK01073280 AEFK01073281 AEFK01073282 ERE91089.1 AK148105 BAE28346.1 KK719754 KFO66065.1 QUSF01000052 RLV97818.1
KQ460685 KPJ12859.1 KQ459072 KPJ03858.1 GBYB01004832 JAG74599.1 KK853016 KDR12475.1 KQ981805 KYN35548.1 NNAY01005719 OXU16575.1 AAZX01012632 AAZX01017245 GL762073 EFZ22294.1 KQ976403 KYM92136.1 ADTU01002059 KQ978801 KYN28297.1 NEVH01026389 PNF14346.1 PNF14343.1 GBYB01004827 JAG74594.1 GL888531 EGI59668.1 KQ978331 KYM95064.1 KQ414631 KOC67275.1 GBYB01004828 JAG74595.1 GECL01001791 JAP04333.1 GEBQ01011011 JAT28966.1 GBGD01000618 JAC88271.1 GFTR01007859 JAW08567.1 GAKP01003115 JAC55837.1 CH478401 EAT33062.1 GAKP01003114 JAC55838.1 GECU01011193 JAS96513.1 GAMC01006366 JAC00190.1 QOIP01000005 RLU22676.1 JXUM01029787 JXUM01029788 KQ560864 KXJ80582.1 KQ435711 KOX79647.1 GAPW01000357 JAC13241.1 UFQS01000627 UFQT01000627 SSX05525.1 SSX25884.1 GECZ01012666 JAS57103.1 GBBI01004796 JAC13916.1 AAAB01008846 EAA06509.5 GEDC01004195 JAS33103.1 AJVK01017844 AJVK01017845 APCN01001316 GFDL01009836 JAV25209.1 GGFM01003083 MBW23834.1 GGFM01003078 MBW23829.1 GFDL01009263 JAV25782.1 ATLV01014931 KE524996 KFB39651.1 GFDL01009553 JAV25492.1 KZ288289 PBC29266.1 GGFJ01002703 MBW51844.1 DS232933 EDS26872.1 GDKW01000077 JAI56518.1 AYCK01002591 AYCK01002592 AYCK01002593 GBXI01008103 JAD06189.1 BC044110 BC108785 CM004467 AAH44110.1 AAI08786.1 OCT98127.1 KE162863 EPQ09913.1 AEMK02000096 DQIR01199597 HDB55074.1 JXUM01012471 KQ560355 KXJ82864.1 AAPE02038420 AAGJ04021643 KE663652 ERE91092.1 AAEX03014792 JH000563 EGW12303.1 ERE91090.1 AEYP01066060 GABZ01004591 JAA48934.1 AK048876 AK051387 AL606521 BC013537 BC024840 GFDF01008307 JAV05777.1 ERE91091.1 BT020851 AAX08868.1 AANG04000189 AEFK01073279 AEFK01073280 AEFK01073281 AEFK01073282 ERE91089.1 AK148105 BAE28346.1 KK719754 KFO66065.1 QUSF01000052 RLV97818.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000027135
+ More
UP000078541 UP000215335 UP000002358 UP000078540 UP000005205 UP000078492 UP000235965 UP000007755 UP000078542 UP000053825 UP000075880 UP000008820 UP000279307 UP000069940 UP000249989 UP000053105 UP000007062 UP000092462 UP000075840 UP000005203 UP000075903 UP000076407 UP000030765 UP000242457 UP000002320 UP000075902 UP000079721 UP000075885 UP000095300 UP000028760 UP000245340 UP000186698 UP000248481 UP000008227 UP000001074 UP000002281 UP000007110 UP000030759 UP000002254 UP000001075 UP000000715 UP000000589 UP000248482 UP000007646 UP000002280 UP000011712 UP000007648 UP000245341 UP000189706 UP000233120 UP000001811 UP000052976 UP000233020 UP000276834 UP000248480
UP000078541 UP000215335 UP000002358 UP000078540 UP000005205 UP000078492 UP000235965 UP000007755 UP000078542 UP000053825 UP000075880 UP000008820 UP000279307 UP000069940 UP000249989 UP000053105 UP000007062 UP000092462 UP000075840 UP000005203 UP000075903 UP000076407 UP000030765 UP000242457 UP000002320 UP000075902 UP000079721 UP000075885 UP000095300 UP000028760 UP000245340 UP000186698 UP000248481 UP000008227 UP000001074 UP000002281 UP000007110 UP000030759 UP000002254 UP000001075 UP000000715 UP000000589 UP000248482 UP000007646 UP000002280 UP000011712 UP000007648 UP000245341 UP000189706 UP000233120 UP000001811 UP000052976 UP000233020 UP000276834 UP000248480
Pfam
PF02845 CUE
Interpro
ProteinModelPortal
H9JCU9
A0A212F0K6
A0A2A4JAH9
A0A194R5R7
A0A0N0P9X2
A0A0C9QW05
+ More
A0A067R101 A0A195F513 A0A232EE01 K7JAM1 E9IAW0 A0A195BVZ7 A0A158NRK5 A0A195EK54 A0A2J7PDC7 A0A2J7PDE4 A0A0C9QVZ9 F4X1L0 A0A151I8S3 A0A0L7R8S9 A0A0C9PU20 A0A0V0G8Y4 A0A182JD78 A0A1B6LZ56 A0A069DWR0 A0A224X864 A0A034WNB7 Q16FQ3 A0A034WQ05 A0A1B6JBW3 W8C9T6 A0A3L8DQW0 A0A182H8Y8 A0A0M9A8Y7 A0A023EVX1 A0A336KQ35 A0A1B6G460 A0A023EY09 Q7QFD7 A0A1B6E5A1 A0A1B0GQR2 A0A182IAT0 A0A088AR41 A0A182UVX2 A0A1Q3FCG2 A0A182XDS2 A0A2M3Z5Q5 A0A2M3Z5N1 A0A1Q3FE69 A0A084VNV7 A0A1Q3FDB6 A0A2A3EDL8 A0A2M4BFN4 B0XFK9 A0A182U305 A0A0P4VUW3 A0A1S3WJ36 A0A182PBK8 A0A1I8NME4 A0A096LYT5 A0A2U3VX67 A0A0A1X4E1 Q7ZXU8 S7PH01 A0A2Y9I5L4 A0A286ZQL9 A0A182GIZ2 G1PWT1 F6U4G2 W4YDX4 A0A061INR1 E2RS73 G3HP16 M3YP83 K9J2P7 Q91WR3 A0A2Y9L4D5 A0A1L8DHC1 A0A061IMU8 G3TQL6 Q91WR3-3 Q5E9R9 F7B2Z2 M3W1V2 G3VLN4 A0A2U3Y7Z7 A0A3Q0CNE6 A0A2Y9I5D8 A0A2K6BI62 A0A061IQW0 G1U1Z1 Q3UG63 A0A091FXE8 A0A2K5F1L4 A0A3L8S7C0 A0A2Y9RD40
A0A067R101 A0A195F513 A0A232EE01 K7JAM1 E9IAW0 A0A195BVZ7 A0A158NRK5 A0A195EK54 A0A2J7PDC7 A0A2J7PDE4 A0A0C9QVZ9 F4X1L0 A0A151I8S3 A0A0L7R8S9 A0A0C9PU20 A0A0V0G8Y4 A0A182JD78 A0A1B6LZ56 A0A069DWR0 A0A224X864 A0A034WNB7 Q16FQ3 A0A034WQ05 A0A1B6JBW3 W8C9T6 A0A3L8DQW0 A0A182H8Y8 A0A0M9A8Y7 A0A023EVX1 A0A336KQ35 A0A1B6G460 A0A023EY09 Q7QFD7 A0A1B6E5A1 A0A1B0GQR2 A0A182IAT0 A0A088AR41 A0A182UVX2 A0A1Q3FCG2 A0A182XDS2 A0A2M3Z5Q5 A0A2M3Z5N1 A0A1Q3FE69 A0A084VNV7 A0A1Q3FDB6 A0A2A3EDL8 A0A2M4BFN4 B0XFK9 A0A182U305 A0A0P4VUW3 A0A1S3WJ36 A0A182PBK8 A0A1I8NME4 A0A096LYT5 A0A2U3VX67 A0A0A1X4E1 Q7ZXU8 S7PH01 A0A2Y9I5L4 A0A286ZQL9 A0A182GIZ2 G1PWT1 F6U4G2 W4YDX4 A0A061INR1 E2RS73 G3HP16 M3YP83 K9J2P7 Q91WR3 A0A2Y9L4D5 A0A1L8DHC1 A0A061IMU8 G3TQL6 Q91WR3-3 Q5E9R9 F7B2Z2 M3W1V2 G3VLN4 A0A2U3Y7Z7 A0A3Q0CNE6 A0A2Y9I5D8 A0A2K6BI62 A0A061IQW0 G1U1Z1 Q3UG63 A0A091FXE8 A0A2K5F1L4 A0A3L8S7C0 A0A2Y9RD40
PDB
2DI0
E-value=1.52997e-10,
Score=161
Ontologies
GO
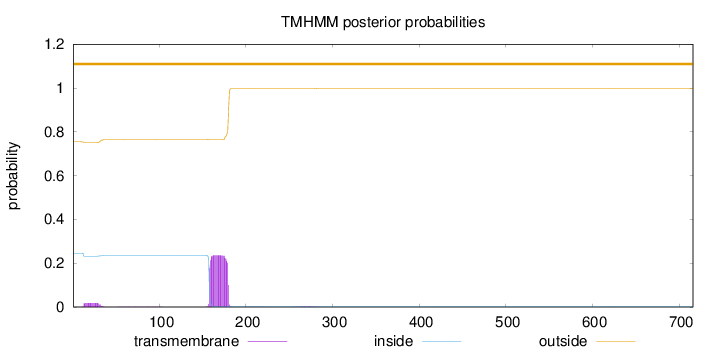
Topology
Subcellular location
Nucleus
Colocalizes with the spliceosomal components PRPF8 and SNRNP200/BRR2 in nuclear foci when cells have been exposed to alkylating agents that cause DNA damage. Colocalizes with RNF113A and 'Lys-63'-linked polyubiquitinated proteins, ALKBH3 and ASCC3 in nuclear foci when cells have been exposed to alkylating agents that cause DNA damage. With evidence from 1 publications.
Nucleus speckle Colocalizes with the spliceosomal components PRPF8 and SNRNP200/BRR2 in nuclear foci when cells have been exposed to alkylating agents that cause DNA damage. Colocalizes with RNF113A and 'Lys-63'-linked polyubiquitinated proteins, ALKBH3 and ASCC3 in nuclear foci when cells have been exposed to alkylating agents that cause DNA damage. With evidence from 1 publications.
Nucleus speckle Colocalizes with the spliceosomal components PRPF8 and SNRNP200/BRR2 in nuclear foci when cells have been exposed to alkylating agents that cause DNA damage. Colocalizes with RNF113A and 'Lys-63'-linked polyubiquitinated proteins, ALKBH3 and ASCC3 in nuclear foci when cells have been exposed to alkylating agents that cause DNA damage. With evidence from 1 publications.
Length:
716
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.53987000000001
Exp number, first 60 AAs:
0.35845
Total prob of N-in:
0.24525
outside
1 - 716
Population Genetic Test Statistics
Pi
196.202578
Theta
32.176886
Tajima's D
0.652373
CLR
3.032503
CSRT
0.569021548922554
Interpretation
Uncertain
