Gene
KWMTBOMO01082
Pre Gene Modal
BGIBMGA007347
Annotation
olfactory_receptor_39_[Bombyx_mori]
Full name
Odorant receptor
Location in the cell
PlasmaMembrane Reliability : 4.382
Sequence
CDS
ATGCAGTCCGTTAAGAAGCGCGGCGTAATTTTCTGGCTAGTGATAATTAGTAATGGAGTTGTCTACCTCGTGAAGCCAATCGTGACGCCCGGCAGGCATTTCATGGAAGACCAATTCATCATTTTAGGTCTCGAACCTAAGTACGAAACACCGAATTACGAGATAGGATTTTTTATGATGGCGGTCGGAGTTTGTGTGACTTGCTACTTGCCAGCCAACATCACGGCTTACCTGATAACCGTAGCCGGATATTCAGAAGCACAGTTTTTGGCACTTGGACACGAACTCGCAAACCTGTGGCCGGACGCTCAACTACACTGCCGCGCAATGAACTTGAGCCAATCAGTAAATGAGCAAGCTAACGAATACGTGAAAATGCGATTGAGAGAATTAGTAAAAATACATTCCACCAATGTTAACCTTTTGAGGGATATAGAGGGAGCTTTTAGAGGAGCTATTGCTGTTGAATTTTTACTATTGATTGTGGGCCTTATAGCTGAGTTGCTGGGGGGCTTGGAGAATACATACATGCAGGTGCCTTTTGCGCTGATTCAGGTCTCGGTGGACTGCCTCACGGGACAGCGGGTGATGGATGCGAACTTGGCTCTAGAACGCGCAGTTTACGATTGTCGGTGGGAAGAGTTCGATGCTAGTAACCGTCGGGTCGTGTTGCTACTGTTGCAGAACGCACAGAAAGTAGCCACGCTGTCGGCGGGCGGCATCGCCACCCTCAACTTCAGCTGTCTCATGGCTGTGATCAAATCTATCTACTCTGCCTACACCACGCTTAGAACTACCATGAAATAA
Protein
MQSVKKRGVIFWLVIISNGVVYLVKPIVTPGRHFMEDQFIILGLEPKYETPNYEIGFFMMAVGVCVTCYLPANITAYLITVAGYSEAQFLALGHELANLWPDAQLHCRAMNLSQSVNEQANEYVKMRLRELVKIHSTNVNLLRDIEGAFRGAIAVEFLLLIVGLIAELLGGLENTYMQVPFALIQVSVDCLTGQRVMDANLALERAVYDCRWEEFDASNRRVVLLLLQNAQKVATLSAGGIATLNFSCLMAVIKSIYSAYTTLRTTMK
Summary
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Uniprot
B1B1Q7
H9JCV0
A0A0L7LGZ8
A0A0E4B595
A0A1Y9TJT9
A0A0E4B599
+ More
B1B1Q1 A7E3H6 A0A2H1WCF2 A0A3G6V6V3 H9JCZ2 C4B7W9 I3Y3H8 A0A345BEW0 A7E3H9 A0A2W1BR24 A0A076E626 A0A076E5Z5 A0A0V0J1D8 A0A1V1WC68 A0A2A4J488 A0A2L1TG79 A0A2W1BGJ8 A0A0S1TPK5 A0A1S5XXM9 A0A2L1TG85 A0A1V1WC56 A0A1S5XXN2 A0A2K8GL41 A0A1B3B749 A0A0K8TVD3 A0A146JVP7 A0A1S5XXN8 A0A3G6V712 A0A2H1WDS1 A0A0L7K4E6 A0A1Y9TJJ5 A0A146JWS3 A0A2A4J308 A0A075T2Z3 A0A0B5CSS9 A0A2W1BIS7 A0A0B5GQ12 A0A2L1TG44 A0A1X9P7J6 A0A097ITU7 A0A097IYM0 A0A146JYU4 A0A0K8TU65 A0A0B5A1T0 A0A2A4JBA0 A0A097IYN8 A0A0B5GR64 A0A0B5CUS1 A0A097ITT9 A0A2H1WC45 A0A076E9A9 A0A2K8GKT9 A0A076E995 A0A2A4JAT4 H9A5Q5 A0A2H1WC18 A0A345BEX0 A7E3H8 C4B7W8 A0A0S1TRB2 A0A1B3B753 A0A2W1BH90 A0A1S5XXN3 A0A2L1TG55 A0A2A4JA14 A0A1B3P5T0 H9A5Q6 A0A223HDE2 A2T872 A0A345BEX3 A0A2K8GKS3 A0A223HD72 A0A1S5XXN5 A0A223HD20 A0A1B2G2M2 A0A223HD73 A0A212FKR6 A0A0E4B423 A0A212ERN2 A0A386H9N9 A0A212FC05 A0A212FC10 H9J5G9 I3Y3H7 A0A212FC17 A0A0U2RFI8 A0A345BEV9
B1B1Q1 A7E3H6 A0A2H1WCF2 A0A3G6V6V3 H9JCZ2 C4B7W9 I3Y3H8 A0A345BEW0 A7E3H9 A0A2W1BR24 A0A076E626 A0A076E5Z5 A0A0V0J1D8 A0A1V1WC68 A0A2A4J488 A0A2L1TG79 A0A2W1BGJ8 A0A0S1TPK5 A0A1S5XXM9 A0A2L1TG85 A0A1V1WC56 A0A1S5XXN2 A0A2K8GL41 A0A1B3B749 A0A0K8TVD3 A0A146JVP7 A0A1S5XXN8 A0A3G6V712 A0A2H1WDS1 A0A0L7K4E6 A0A1Y9TJJ5 A0A146JWS3 A0A2A4J308 A0A075T2Z3 A0A0B5CSS9 A0A2W1BIS7 A0A0B5GQ12 A0A2L1TG44 A0A1X9P7J6 A0A097ITU7 A0A097IYM0 A0A146JYU4 A0A0K8TU65 A0A0B5A1T0 A0A2A4JBA0 A0A097IYN8 A0A0B5GR64 A0A0B5CUS1 A0A097ITT9 A0A2H1WC45 A0A076E9A9 A0A2K8GKT9 A0A076E995 A0A2A4JAT4 H9A5Q5 A0A2H1WC18 A0A345BEX0 A7E3H8 C4B7W8 A0A0S1TRB2 A0A1B3B753 A0A2W1BH90 A0A1S5XXN3 A0A2L1TG55 A0A2A4JA14 A0A1B3P5T0 H9A5Q6 A0A223HDE2 A2T872 A0A345BEX3 A0A2K8GKS3 A0A223HD72 A0A1S5XXN5 A0A223HD20 A0A1B2G2M2 A0A223HD73 A0A212FKR6 A0A0E4B423 A0A212ERN2 A0A386H9N9 A0A212FC05 A0A212FC10 H9J5G9 I3Y3H7 A0A212FC17 A0A0U2RFI8 A0A345BEV9
Pubmed
EMBL
AB186516
AB472118
BAG12815.1
BAH66336.1
BABH01027717
BABH01027718
+ More
BABH01027719 BABH01027720 BABH01027721 JTDY01001167 KOB74670.1 LC002703 BAR43451.1 KX084505 ARO76460.1 LC002710 BAR43458.1 AB186510 AB472114 BAG12809.1 BAH66332.1 BK005941 DAA05991.1 ODYU01007660 SOQ50636.1 MH193989 AZB49435.1 BABH01027593 AB472117 BAH66335.1 JQ794808 AFL70813.1 MG816599 AXF48784.1 BK005944 DAA05994.1 KZ150062 PZC74193.1 KF487708 AII01106.1 KF487678 AII01076.1 GDKB01000050 JAP38446.1 GENK01000073 JAV45840.1 NWSH01003458 PCG66260.1 KY707234 AVF19676.1 PZC74192.1 KR935736 ALM26228.1 KY399281 AQQ73512.1 KY707208 AVF19650.1 GENK01000057 JAV45856.1 KY399282 AQQ73513.1 KY225496 ARO70508.1 KT588132 AOE48042.1 GCVX01000195 JAI18035.1 GEDO01000034 JAP88592.1 KY399283 AQQ73514.1 MH193983 AZB49429.1 SOQ50634.1 JTDY01012011 KOB52347.1 KX084500 ARO76455.1 GEDO01000058 JAP88568.1 PCG66259.1 KF768704 AIG51898.1 KM678313 AJE25886.1 PZC74191.1 KM892387 AJF23802.1 KY707206 AVF19648.1 KX084467 ARO76422.1 KM597213 AIT69890.1 KM655550 AIT72000.1 GEDO01000057 JAP88569.1 GCVX01000196 JAI18034.1 KJ542684 AJD81568.1 NWSH01002174 PCG68968.1 KM655551 AIT72001.1 KM892388 AJF23803.1 KM678314 AJE25887.1 KM597214 AIT69891.1 SOQ50633.1 KF487680 AII01078.1 KX585338 ARO70231.1 KF487666 AII01064.1 PCG68969.1 JN836705 AFC91745.2 SOQ50635.1 MG816609 AXF48794.1 BK005943 DAA05993.1 AB472116 BAH66334.1 KR935735 ALM26227.1 KT588137 AOE48047.1 PZC74198.1 KY399285 AQQ73516.1 KY707207 AVF19649.1 PCG68967.1 KX655993 AOG12942.1 JN836706 AFC91746.1 KY283740 AST36399.1 DQ991156 ABK27850.1 MG816612 AXF48797.1 KX585345 ARO70238.1 KY283662 AST36321.1 KY399284 AQQ73515.1 KY283620 AST36279.1 KU598896 ANZ03142.1 KY283663 AST36322.1 AGBW02008001 OWR54326.1 LC002711 BAR43459.1 AGBW02012963 OWR44153.1 MG546606 AYD42229.1 AGBW02009241 OWR51276.1 OWR51272.1 BABH01028070 BABH01028071 BABH01028072 BABH01028073 JQ794807 AFL70812.1 OWR51275.1 KP975144 ALT31663.1 MG816598 AXF48783.1
BABH01027719 BABH01027720 BABH01027721 JTDY01001167 KOB74670.1 LC002703 BAR43451.1 KX084505 ARO76460.1 LC002710 BAR43458.1 AB186510 AB472114 BAG12809.1 BAH66332.1 BK005941 DAA05991.1 ODYU01007660 SOQ50636.1 MH193989 AZB49435.1 BABH01027593 AB472117 BAH66335.1 JQ794808 AFL70813.1 MG816599 AXF48784.1 BK005944 DAA05994.1 KZ150062 PZC74193.1 KF487708 AII01106.1 KF487678 AII01076.1 GDKB01000050 JAP38446.1 GENK01000073 JAV45840.1 NWSH01003458 PCG66260.1 KY707234 AVF19676.1 PZC74192.1 KR935736 ALM26228.1 KY399281 AQQ73512.1 KY707208 AVF19650.1 GENK01000057 JAV45856.1 KY399282 AQQ73513.1 KY225496 ARO70508.1 KT588132 AOE48042.1 GCVX01000195 JAI18035.1 GEDO01000034 JAP88592.1 KY399283 AQQ73514.1 MH193983 AZB49429.1 SOQ50634.1 JTDY01012011 KOB52347.1 KX084500 ARO76455.1 GEDO01000058 JAP88568.1 PCG66259.1 KF768704 AIG51898.1 KM678313 AJE25886.1 PZC74191.1 KM892387 AJF23802.1 KY707206 AVF19648.1 KX084467 ARO76422.1 KM597213 AIT69890.1 KM655550 AIT72000.1 GEDO01000057 JAP88569.1 GCVX01000196 JAI18034.1 KJ542684 AJD81568.1 NWSH01002174 PCG68968.1 KM655551 AIT72001.1 KM892388 AJF23803.1 KM678314 AJE25887.1 KM597214 AIT69891.1 SOQ50633.1 KF487680 AII01078.1 KX585338 ARO70231.1 KF487666 AII01064.1 PCG68969.1 JN836705 AFC91745.2 SOQ50635.1 MG816609 AXF48794.1 BK005943 DAA05993.1 AB472116 BAH66334.1 KR935735 ALM26227.1 KT588137 AOE48047.1 PZC74198.1 KY399285 AQQ73516.1 KY707207 AVF19649.1 PCG68967.1 KX655993 AOG12942.1 JN836706 AFC91746.1 KY283740 AST36399.1 DQ991156 ABK27850.1 MG816612 AXF48797.1 KX585345 ARO70238.1 KY283662 AST36321.1 KY399284 AQQ73515.1 KY283620 AST36279.1 KU598896 ANZ03142.1 KY283663 AST36322.1 AGBW02008001 OWR54326.1 LC002711 BAR43459.1 AGBW02012963 OWR44153.1 MG546606 AYD42229.1 AGBW02009241 OWR51276.1 OWR51272.1 BABH01028070 BABH01028071 BABH01028072 BABH01028073 JQ794807 AFL70812.1 OWR51275.1 KP975144 ALT31663.1 MG816598 AXF48783.1
Proteomes
Pfam
PF02949 7tm_6
Interpro
IPR004117
7tm6_olfct_rcpt
ProteinModelPortal
B1B1Q7
H9JCV0
A0A0L7LGZ8
A0A0E4B595
A0A1Y9TJT9
A0A0E4B599
+ More
B1B1Q1 A7E3H6 A0A2H1WCF2 A0A3G6V6V3 H9JCZ2 C4B7W9 I3Y3H8 A0A345BEW0 A7E3H9 A0A2W1BR24 A0A076E626 A0A076E5Z5 A0A0V0J1D8 A0A1V1WC68 A0A2A4J488 A0A2L1TG79 A0A2W1BGJ8 A0A0S1TPK5 A0A1S5XXM9 A0A2L1TG85 A0A1V1WC56 A0A1S5XXN2 A0A2K8GL41 A0A1B3B749 A0A0K8TVD3 A0A146JVP7 A0A1S5XXN8 A0A3G6V712 A0A2H1WDS1 A0A0L7K4E6 A0A1Y9TJJ5 A0A146JWS3 A0A2A4J308 A0A075T2Z3 A0A0B5CSS9 A0A2W1BIS7 A0A0B5GQ12 A0A2L1TG44 A0A1X9P7J6 A0A097ITU7 A0A097IYM0 A0A146JYU4 A0A0K8TU65 A0A0B5A1T0 A0A2A4JBA0 A0A097IYN8 A0A0B5GR64 A0A0B5CUS1 A0A097ITT9 A0A2H1WC45 A0A076E9A9 A0A2K8GKT9 A0A076E995 A0A2A4JAT4 H9A5Q5 A0A2H1WC18 A0A345BEX0 A7E3H8 C4B7W8 A0A0S1TRB2 A0A1B3B753 A0A2W1BH90 A0A1S5XXN3 A0A2L1TG55 A0A2A4JA14 A0A1B3P5T0 H9A5Q6 A0A223HDE2 A2T872 A0A345BEX3 A0A2K8GKS3 A0A223HD72 A0A1S5XXN5 A0A223HD20 A0A1B2G2M2 A0A223HD73 A0A212FKR6 A0A0E4B423 A0A212ERN2 A0A386H9N9 A0A212FC05 A0A212FC10 H9J5G9 I3Y3H7 A0A212FC17 A0A0U2RFI8 A0A345BEV9
B1B1Q1 A7E3H6 A0A2H1WCF2 A0A3G6V6V3 H9JCZ2 C4B7W9 I3Y3H8 A0A345BEW0 A7E3H9 A0A2W1BR24 A0A076E626 A0A076E5Z5 A0A0V0J1D8 A0A1V1WC68 A0A2A4J488 A0A2L1TG79 A0A2W1BGJ8 A0A0S1TPK5 A0A1S5XXM9 A0A2L1TG85 A0A1V1WC56 A0A1S5XXN2 A0A2K8GL41 A0A1B3B749 A0A0K8TVD3 A0A146JVP7 A0A1S5XXN8 A0A3G6V712 A0A2H1WDS1 A0A0L7K4E6 A0A1Y9TJJ5 A0A146JWS3 A0A2A4J308 A0A075T2Z3 A0A0B5CSS9 A0A2W1BIS7 A0A0B5GQ12 A0A2L1TG44 A0A1X9P7J6 A0A097ITU7 A0A097IYM0 A0A146JYU4 A0A0K8TU65 A0A0B5A1T0 A0A2A4JBA0 A0A097IYN8 A0A0B5GR64 A0A0B5CUS1 A0A097ITT9 A0A2H1WC45 A0A076E9A9 A0A2K8GKT9 A0A076E995 A0A2A4JAT4 H9A5Q5 A0A2H1WC18 A0A345BEX0 A7E3H8 C4B7W8 A0A0S1TRB2 A0A1B3B753 A0A2W1BH90 A0A1S5XXN3 A0A2L1TG55 A0A2A4JA14 A0A1B3P5T0 H9A5Q6 A0A223HDE2 A2T872 A0A345BEX3 A0A2K8GKS3 A0A223HD72 A0A1S5XXN5 A0A223HD20 A0A1B2G2M2 A0A223HD73 A0A212FKR6 A0A0E4B423 A0A212ERN2 A0A386H9N9 A0A212FC05 A0A212FC10 H9J5G9 I3Y3H7 A0A212FC17 A0A0U2RFI8 A0A345BEV9
PDB
6C70
E-value=0.00610157,
Score=91
Ontologies
KEGG
GO
PANTHER
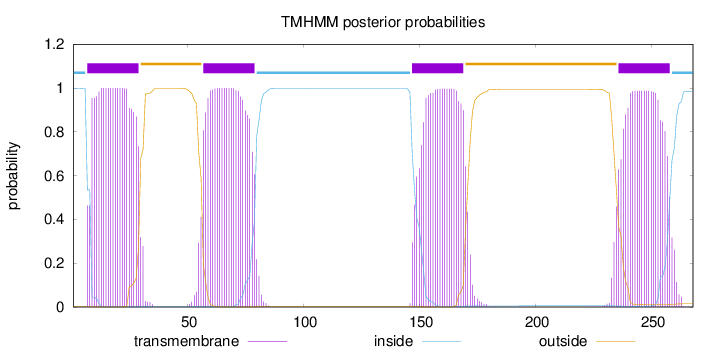
Topology
Subcellular location
Cell membrane
Length:
268
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
88.4253
Exp number, first 60 AAs:
26.26359
Total prob of N-in:
0.99829
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 56
TMhelix
57 - 79
inside
80 - 146
TMhelix
147 - 169
outside
170 - 235
TMhelix
236 - 258
inside
259 - 268
Population Genetic Test Statistics
Pi
206.699383
Theta
147.712681
Tajima's D
-0.585312
CLR
0.244949
CSRT
0.219789010549473
Interpretation
Uncertain
