Gene
KWMTBOMO01076 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007418
Annotation
adenosine_kinase_[Bombyx_mori]
Full name
Receptor protein serine/threonine kinase
Location in the cell
Cytoplasmic Reliability : 1.413 Extracellular Reliability : 1.841
Sequence
CDS
ATGGACGTTTCTGATTCCATATGTGACCAAGAAGGATTATTAGTGGGCATAGGGAATCCTTTGTTGGACATATCCGCCAGTGTTGACGAGGACTTGCTGAAGAAGTATGACCTGCACCCCGACGATGCCATTATGGCTGAAGAAAAACACATGCCACTCTACTCGGAACTCGTTGACAAGTATAATGCAGAGTATATTGCTGGTGGAAGCGTTCAGAATTCATTAAGAGTAGCACAATGGATTCTAAAGAAACCAAATATTTGTACTTACTTTGGCTGTGTGGGAAATGATGAATATGCTAAATTATTAAAGGAGAGAGCAATTGCAGATGGAGTCCACGTTCAATATCAAGTATCAAATGAGGTAGCAACTGGTACATGTGCAGTGCTAGTAACTGGTACACACCGCTCACTGTGCGCCAATCTCGGTGCTGCACAACACTTCACACCAGATCATTTACAGAAAGAAGAATGCAAGAAAAGCATTGAAGCAGCGAAGTTCTTTTATGCTTCTGGTTTCTTTGTGGCAGTTTCTCCAGAGTCGATATTGTTATTGGCCCAGCATGCGCACGATAATGGACATACGTTTGTTATGAACTTGAGTGCGCCCTTTGTCTCACAGTTCTATAAGGAACCGCTCGAGAAACTTCTCCCATATGTCGACGTGCTCTTCGGGAATGAATCGGAGGCAGACGCTTTCGCGAAGGCATTCAACATAAATTCGAGCGATGTGCAGGAGATCGCTTTACGGATCGCCAGCATGCCCAAATTGAACGCGAACCGTCAGCGCGTCGTGGTCATCACGCAGGGCTGCCAACCCGTCGTGCTGGTACAGAGCGGACGAGTCACCCTGATACCGGTCGAGGCTTTGCCGAGGGAACGTATCATTGACACAAACGGTGCCGGAGATGCCTTTACTGGTGGGTACTTGGCGCAGCTCGTGCTGAACCGCGAGCCAGCCGCCTGCGTGCGGTGCGCTGTCTACTGTGCCACGCATGTCATCCAGCACCCGGGCTGCACATTCAGCGGACCCAGCGAATACAATGACTGA
Protein
MDVSDSICDQEGLLVGIGNPLLDISASVDEDLLKKYDLHPDDAIMAEEKHMPLYSELVDKYNAEYIAGGSVQNSLRVAQWILKKPNICTYFGCVGNDEYAKLLKERAIADGVHVQYQVSNEVATGTCAVLVTGTHRSLCANLGAAQHFTPDHLQKEECKKSIEAAKFFYASGFFVAVSPESILLLAQHAHDNGHTFVMNLSAPFVSQFYKEPLEKLLPYVDVLFGNESEADAFAKAFNINSSDVQEIALRIASMPKLNANRQRVVVITQGCQPVVLVQSGRVTLIPVEALPRERIIDTNGAGDAFTGGYLAQLVLNREPAACVRCAVYCATHVIQHPGCTFSGPSEYND
Summary
Catalytic Activity
[receptor-protein]-L-serine + ATP = [receptor-protein]-O-phospho-L-serine + ADP + H(+)
[receptor-protein]-L-threonine + ATP = [receptor-protein]-O-phospho-L-threonine + ADP + H(+)
[receptor-protein]-L-threonine + ATP = [receptor-protein]-O-phospho-L-threonine + ADP + H(+)
Similarity
Belongs to the protein kinase superfamily. TKL Ser/Thr protein kinase family. TGFB receptor subfamily.
Uniprot
Q2F635
A0A194QBY3
A0A1I8PKS1
A0A1I8PKZ2
A0A0L0CEB8
A0A182PBL9
+ More
A0A182V6Y9 A0A182WZC4 A0A182HPX9 A0A182KSU8 A0A1I8MVS8 A0A1I8MVS6 A0A182U6C2 A0A2W1BIT6 A0A182Y1X9 A0A182JWJ2 A0A182SV27 A0A158P348 A0A0J7KXH2 A0A182PZK2 A0A182W9A9 A0A182RGK6 D6WX36 A0A182J1V8 A0A182NPF1 A0A3L8E2Y0 A0A1A9WSS3 A0A1B0A694 A0A1S4EXT2 A0A1Q3FEE8 A0A1B0FA86 D3TS39 A0A182M5Y4 Q1HQJ8 A0A1Q3FDY0 A0A1L8E206 B0WEK9 E9IIK6 A0A1A9VKW1 F4W4H8 A0A1B0AWD3 A0A182GS07 A0A1B0CRL5 A0A1L8E2F0 A0A1A9Y2T2 A0A1L8E1V5 W8C768 A0A084WB55 A0A087ZS94 A0A0A1XI02 A0A1W4VIX3 V9IHF0 A0A0A1XNF6 A0A1W4V6S3 A0A2R7WNX4 A0A232EIL7 A0A067RTD0 A0A336M4V2 B4HGN5 B4L0J3 A0A0K8V7Z8 A0A0K8V3D4 A0A034W2N7 A0A0Q9XD88 A0A034W125 E2A1C3 A0A1W4XE72 A0A2J7R4D5 A0A0K8VX64 B4MM65 A0A0K8W0U2 A0A3B0J432 A0A1Y1LM30 A0A0A9YAB8 N6TS29 A0A1B6J821 B4LBH2 A0A0Q9WTX8 A0A0V0G398 B4GRU4 A0A0L7RD75 A0A023F891 B5DPJ4 A0A161TIT7 A0A1B6DN45 A0A0P8XT85 B4PHB3 B3MAT9 R4G8F0 A0A0P4W088 B4K005 A0A1A9UKT6 A0A1B0FM18 A0A154PIR7 Q86NZ5 A0A0R1DXF0 Q9VU39 Q9VU38 B3NCY4 B4QIY5
A0A182V6Y9 A0A182WZC4 A0A182HPX9 A0A182KSU8 A0A1I8MVS8 A0A1I8MVS6 A0A182U6C2 A0A2W1BIT6 A0A182Y1X9 A0A182JWJ2 A0A182SV27 A0A158P348 A0A0J7KXH2 A0A182PZK2 A0A182W9A9 A0A182RGK6 D6WX36 A0A182J1V8 A0A182NPF1 A0A3L8E2Y0 A0A1A9WSS3 A0A1B0A694 A0A1S4EXT2 A0A1Q3FEE8 A0A1B0FA86 D3TS39 A0A182M5Y4 Q1HQJ8 A0A1Q3FDY0 A0A1L8E206 B0WEK9 E9IIK6 A0A1A9VKW1 F4W4H8 A0A1B0AWD3 A0A182GS07 A0A1B0CRL5 A0A1L8E2F0 A0A1A9Y2T2 A0A1L8E1V5 W8C768 A0A084WB55 A0A087ZS94 A0A0A1XI02 A0A1W4VIX3 V9IHF0 A0A0A1XNF6 A0A1W4V6S3 A0A2R7WNX4 A0A232EIL7 A0A067RTD0 A0A336M4V2 B4HGN5 B4L0J3 A0A0K8V7Z8 A0A0K8V3D4 A0A034W2N7 A0A0Q9XD88 A0A034W125 E2A1C3 A0A1W4XE72 A0A2J7R4D5 A0A0K8VX64 B4MM65 A0A0K8W0U2 A0A3B0J432 A0A1Y1LM30 A0A0A9YAB8 N6TS29 A0A1B6J821 B4LBH2 A0A0Q9WTX8 A0A0V0G398 B4GRU4 A0A0L7RD75 A0A023F891 B5DPJ4 A0A161TIT7 A0A1B6DN45 A0A0P8XT85 B4PHB3 B3MAT9 R4G8F0 A0A0P4W088 B4K005 A0A1A9UKT6 A0A1B0FM18 A0A154PIR7 Q86NZ5 A0A0R1DXF0 Q9VU39 Q9VU38 B3NCY4 B4QIY5
EC Number
2.7.11.30
Pubmed
19121390
26354079
26108605
20966253
25315136
28756777
+ More
25244985 21347285 18362917 19820115 30249741 20353571 17204158 17510324 21282665 21719571 26483478 24495485 24438588 25830018 28648823 24845553 17994087 25348373 20798317 28004739 25401762 26823975 23537049 25474469 15632085 17550304 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249
25244985 21347285 18362917 19820115 30249741 20353571 17204158 17510324 21282665 21719571 26483478 24495485 24438588 25830018 28648823 24845553 17994087 25348373 20798317 28004739 25401762 26823975 23537049 25474469 15632085 17550304 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249
EMBL
BABH01027708
DQ311237
ABD36182.1
KQ459232
KPJ02500.1
JRES01000502
+ More
KNC30591.1 APCN01003283 KZ150062 PZC74201.1 ADTU01007749 LBMM01002325 KMQ94979.1 AXCN02001826 KQ971361 EFA08050.1 QOIP01000001 RLU26348.1 GFDL01009157 JAV25888.1 CCAG010005457 EZ424241 ADD20517.1 AXCM01002231 DQ440446 CH477208 ABF18479.1 EAT47810.1 GFDL01009265 JAV25780.1 GFDF01001398 JAV12686.1 DS231910 EDS45725.1 GL763494 EFZ19582.1 GL887532 EGI70873.1 JXJN01004651 JXUM01083613 KQ563387 KXJ73934.1 AJWK01024924 GFDF01001399 JAV12685.1 GFDF01001397 JAV12687.1 GAMC01003809 JAC02747.1 ATLV01022314 KE525331 KFB47449.1 GBXI01003747 JAD10545.1 JR047701 AEY60485.1 GBXI01001463 JAD12829.1 KK855177 PTY21294.1 NNAY01004209 OXU18196.1 KK852475 KDR23079.1 UFQT01000391 SSX23889.1 CH480815 EDW41343.1 CH933809 EDW19162.1 GDHF01028455 GDHF01018702 GDHF01017250 JAI23859.1 JAI33612.1 JAI35064.1 GDHF01018900 JAI33414.1 GAKP01010909 JAC48043.1 KRG06523.1 GAKP01010910 JAC48042.1 GL435760 EFN72749.1 NEVH01007425 PNF35701.1 GDHF01008843 JAI43471.1 CH963847 EDW73074.2 GDHF01014834 GDHF01007561 JAI37480.1 JAI44753.1 OUUW01000002 SPP76165.1 GEZM01051782 JAV74702.1 GBHO01015043 GBRD01010871 GBRD01010870 GDHC01001973 JAG28561.1 JAG54953.1 JAQ16656.1 APGK01056679 KB741277 KI209362 KI210050 ENN71091.1 ERL96023.1 ERL96146.1 GECU01012443 JAS95263.1 CH940647 EDW70782.1 KRF85105.1 GECL01003558 JAP02566.1 CH479188 EDW40479.1 KQ414615 KOC68769.1 GBBI01001225 JAC17487.1 CH379069 EDY73542.2 GEMB01000307 JAS02812.1 GEDC01010194 JAS27104.1 CH902618 KPU77928.1 CM000159 EDW94374.1 EDV39173.2 ACPB03002940 GAHY01001087 JAA76423.1 GDKW01001441 JAI55154.1 CH916383 EDV98687.1 CCAG010006681 KQ434931 KZC11776.1 BT003559 AAO39563.1 KRK01798.1 AE014296 AY060709 AAF49852.1 AAL28257.1 AHN58079.1 AAF49853.1 CH954178 EDV51640.1 CM000363 CM002912 EDX10316.1 KMY99350.1
KNC30591.1 APCN01003283 KZ150062 PZC74201.1 ADTU01007749 LBMM01002325 KMQ94979.1 AXCN02001826 KQ971361 EFA08050.1 QOIP01000001 RLU26348.1 GFDL01009157 JAV25888.1 CCAG010005457 EZ424241 ADD20517.1 AXCM01002231 DQ440446 CH477208 ABF18479.1 EAT47810.1 GFDL01009265 JAV25780.1 GFDF01001398 JAV12686.1 DS231910 EDS45725.1 GL763494 EFZ19582.1 GL887532 EGI70873.1 JXJN01004651 JXUM01083613 KQ563387 KXJ73934.1 AJWK01024924 GFDF01001399 JAV12685.1 GFDF01001397 JAV12687.1 GAMC01003809 JAC02747.1 ATLV01022314 KE525331 KFB47449.1 GBXI01003747 JAD10545.1 JR047701 AEY60485.1 GBXI01001463 JAD12829.1 KK855177 PTY21294.1 NNAY01004209 OXU18196.1 KK852475 KDR23079.1 UFQT01000391 SSX23889.1 CH480815 EDW41343.1 CH933809 EDW19162.1 GDHF01028455 GDHF01018702 GDHF01017250 JAI23859.1 JAI33612.1 JAI35064.1 GDHF01018900 JAI33414.1 GAKP01010909 JAC48043.1 KRG06523.1 GAKP01010910 JAC48042.1 GL435760 EFN72749.1 NEVH01007425 PNF35701.1 GDHF01008843 JAI43471.1 CH963847 EDW73074.2 GDHF01014834 GDHF01007561 JAI37480.1 JAI44753.1 OUUW01000002 SPP76165.1 GEZM01051782 JAV74702.1 GBHO01015043 GBRD01010871 GBRD01010870 GDHC01001973 JAG28561.1 JAG54953.1 JAQ16656.1 APGK01056679 KB741277 KI209362 KI210050 ENN71091.1 ERL96023.1 ERL96146.1 GECU01012443 JAS95263.1 CH940647 EDW70782.1 KRF85105.1 GECL01003558 JAP02566.1 CH479188 EDW40479.1 KQ414615 KOC68769.1 GBBI01001225 JAC17487.1 CH379069 EDY73542.2 GEMB01000307 JAS02812.1 GEDC01010194 JAS27104.1 CH902618 KPU77928.1 CM000159 EDW94374.1 EDV39173.2 ACPB03002940 GAHY01001087 JAA76423.1 GDKW01001441 JAI55154.1 CH916383 EDV98687.1 CCAG010006681 KQ434931 KZC11776.1 BT003559 AAO39563.1 KRK01798.1 AE014296 AY060709 AAF49852.1 AAL28257.1 AHN58079.1 AAF49853.1 CH954178 EDV51640.1 CM000363 CM002912 EDX10316.1 KMY99350.1
Proteomes
UP000005204
UP000053268
UP000095300
UP000037069
UP000075885
UP000075903
+ More
UP000076407 UP000075840 UP000075882 UP000095301 UP000075902 UP000076408 UP000075881 UP000075901 UP000005205 UP000036403 UP000075886 UP000075920 UP000075900 UP000007266 UP000075880 UP000075884 UP000279307 UP000091820 UP000092445 UP000092444 UP000075883 UP000008820 UP000002320 UP000078200 UP000007755 UP000092460 UP000069940 UP000249989 UP000092461 UP000092443 UP000030765 UP000005203 UP000192221 UP000215335 UP000027135 UP000001292 UP000009192 UP000000311 UP000192223 UP000235965 UP000007798 UP000268350 UP000019118 UP000030742 UP000008792 UP000008744 UP000053825 UP000001819 UP000007801 UP000002282 UP000015103 UP000001070 UP000076502 UP000000803 UP000008711 UP000000304
UP000076407 UP000075840 UP000075882 UP000095301 UP000075902 UP000076408 UP000075881 UP000075901 UP000005205 UP000036403 UP000075886 UP000075920 UP000075900 UP000007266 UP000075880 UP000075884 UP000279307 UP000091820 UP000092445 UP000092444 UP000075883 UP000008820 UP000002320 UP000078200 UP000007755 UP000092460 UP000069940 UP000249989 UP000092461 UP000092443 UP000030765 UP000005203 UP000192221 UP000215335 UP000027135 UP000001292 UP000009192 UP000000311 UP000192223 UP000235965 UP000007798 UP000268350 UP000019118 UP000030742 UP000008792 UP000008744 UP000053825 UP000001819 UP000007801 UP000002282 UP000015103 UP000001070 UP000076502 UP000000803 UP000008711 UP000000304
Pfam
Interpro
IPR002173
Carboh/pur_kinase_PfkB_CS
+ More
IPR011611 PfkB_dom
IPR001805 Adenokinase
IPR029056 Ribokinase-like
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR001878 Znf_CCHC
IPR003605 GS_dom
IPR000472 Activin_recp
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000333 TGFB_receptor
IPR017441 Protein_kinase_ATP_BS
IPR011611 PfkB_dom
IPR001805 Adenokinase
IPR029056 Ribokinase-like
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR001878 Znf_CCHC
IPR003605 GS_dom
IPR000472 Activin_recp
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000333 TGFB_receptor
IPR017441 Protein_kinase_ATP_BS
Gene 3D
ProteinModelPortal
Q2F635
A0A194QBY3
A0A1I8PKS1
A0A1I8PKZ2
A0A0L0CEB8
A0A182PBL9
+ More
A0A182V6Y9 A0A182WZC4 A0A182HPX9 A0A182KSU8 A0A1I8MVS8 A0A1I8MVS6 A0A182U6C2 A0A2W1BIT6 A0A182Y1X9 A0A182JWJ2 A0A182SV27 A0A158P348 A0A0J7KXH2 A0A182PZK2 A0A182W9A9 A0A182RGK6 D6WX36 A0A182J1V8 A0A182NPF1 A0A3L8E2Y0 A0A1A9WSS3 A0A1B0A694 A0A1S4EXT2 A0A1Q3FEE8 A0A1B0FA86 D3TS39 A0A182M5Y4 Q1HQJ8 A0A1Q3FDY0 A0A1L8E206 B0WEK9 E9IIK6 A0A1A9VKW1 F4W4H8 A0A1B0AWD3 A0A182GS07 A0A1B0CRL5 A0A1L8E2F0 A0A1A9Y2T2 A0A1L8E1V5 W8C768 A0A084WB55 A0A087ZS94 A0A0A1XI02 A0A1W4VIX3 V9IHF0 A0A0A1XNF6 A0A1W4V6S3 A0A2R7WNX4 A0A232EIL7 A0A067RTD0 A0A336M4V2 B4HGN5 B4L0J3 A0A0K8V7Z8 A0A0K8V3D4 A0A034W2N7 A0A0Q9XD88 A0A034W125 E2A1C3 A0A1W4XE72 A0A2J7R4D5 A0A0K8VX64 B4MM65 A0A0K8W0U2 A0A3B0J432 A0A1Y1LM30 A0A0A9YAB8 N6TS29 A0A1B6J821 B4LBH2 A0A0Q9WTX8 A0A0V0G398 B4GRU4 A0A0L7RD75 A0A023F891 B5DPJ4 A0A161TIT7 A0A1B6DN45 A0A0P8XT85 B4PHB3 B3MAT9 R4G8F0 A0A0P4W088 B4K005 A0A1A9UKT6 A0A1B0FM18 A0A154PIR7 Q86NZ5 A0A0R1DXF0 Q9VU39 Q9VU38 B3NCY4 B4QIY5
A0A182V6Y9 A0A182WZC4 A0A182HPX9 A0A182KSU8 A0A1I8MVS8 A0A1I8MVS6 A0A182U6C2 A0A2W1BIT6 A0A182Y1X9 A0A182JWJ2 A0A182SV27 A0A158P348 A0A0J7KXH2 A0A182PZK2 A0A182W9A9 A0A182RGK6 D6WX36 A0A182J1V8 A0A182NPF1 A0A3L8E2Y0 A0A1A9WSS3 A0A1B0A694 A0A1S4EXT2 A0A1Q3FEE8 A0A1B0FA86 D3TS39 A0A182M5Y4 Q1HQJ8 A0A1Q3FDY0 A0A1L8E206 B0WEK9 E9IIK6 A0A1A9VKW1 F4W4H8 A0A1B0AWD3 A0A182GS07 A0A1B0CRL5 A0A1L8E2F0 A0A1A9Y2T2 A0A1L8E1V5 W8C768 A0A084WB55 A0A087ZS94 A0A0A1XI02 A0A1W4VIX3 V9IHF0 A0A0A1XNF6 A0A1W4V6S3 A0A2R7WNX4 A0A232EIL7 A0A067RTD0 A0A336M4V2 B4HGN5 B4L0J3 A0A0K8V7Z8 A0A0K8V3D4 A0A034W2N7 A0A0Q9XD88 A0A034W125 E2A1C3 A0A1W4XE72 A0A2J7R4D5 A0A0K8VX64 B4MM65 A0A0K8W0U2 A0A3B0J432 A0A1Y1LM30 A0A0A9YAB8 N6TS29 A0A1B6J821 B4LBH2 A0A0Q9WTX8 A0A0V0G398 B4GRU4 A0A0L7RD75 A0A023F891 B5DPJ4 A0A161TIT7 A0A1B6DN45 A0A0P8XT85 B4PHB3 B3MAT9 R4G8F0 A0A0P4W088 B4K005 A0A1A9UKT6 A0A1B0FM18 A0A154PIR7 Q86NZ5 A0A0R1DXF0 Q9VU39 Q9VU38 B3NCY4 B4QIY5
PDB
3LOO
E-value=2.7049e-121,
Score=1113
Ontologies
PATHWAY
GO
PANTHER
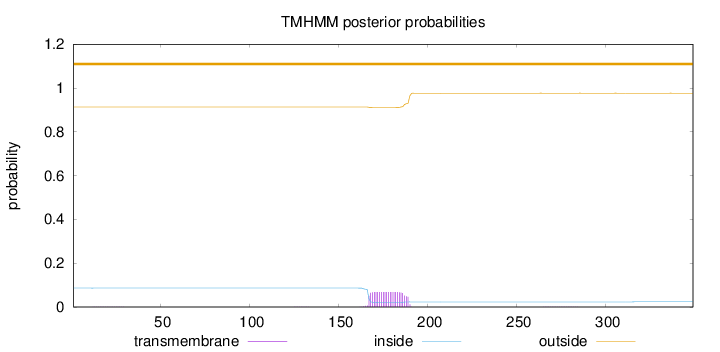
Topology
Length:
349
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.51761
Exp number, first 60 AAs:
0.00559
Total prob of N-in:
0.08664
outside
1 - 349
Population Genetic Test Statistics
Pi
77.64217
Theta
143.476104
Tajima's D
-1.089263
CLR
0.63531
CSRT
0.122293885305735
Interpretation
Uncertain
