Gene
KWMTBOMO01075 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007349
Annotation
heat_shock_protein_60_[Chilo_suppressalis]
Full name
Heat shock protein 60A
Location in the cell
Cytoplasmic Reliability : 1.833 Mitochondrial Reliability : 1.76
Sequence
CDS
ATGAAAGCTCTCATGCTGCAGGGCGTTGACATCCTAGCTGATGCCGTAGCCGTTACAATGGGTCCAAAAGGTAGAAACGTTATTCTGGAACAATCTTGGGGGTCCCCGAAGATCACAAAAGACGGCGTAACAGTCGCCAAAGGTGTTGAACTCAAGGATAAGTTCCAGAACATCGGTGCTAAGTTAGTACAAAACGTAGCTAATAATACTAACGAGGAGGCGGGAGACGGAACTACCACTGCTACAGTACTGGCACGAGCAATCGCGAAGGAGGGCTTCGAAAAAATATCAAAGGGTGCTAACCCGATTGAGATAAGAAGAGGCGTGATGCTCGCCGTAGATGCCGTTAAAGAGAAACTTAAAGGAATGTCGAAACCCGTTACAACACCCGAAGAAATCGCACAAGTAGCTACCATATCTGCTAACGGGGACACGGCAATCGGCAAGCTCATTGCTGATGCAATGAAGAAGGTAGGTAGGGATGGAGTAATCACAGTGAAAGATGGAAAAACCCTCACCGACGAGCTTGAAATCATTGAGGGTATGAAATTTGATAGAGGTTACATTTCACCATACTTCATTAATTCTTCTAAAGGTGCCAAAGTCGAATTCCAGGATGCCCTGGTACTCTTCTCCGAGAAAAAAATTAGCAATGTTCAAACGATCATTCCAGCATTAGAAATGGCTAATCAACAGAGGAAGCCTCTGATCATAGTAGCTGAAGATGTAGACGGAGAGGCACTGTCCACTTTGGTTGTTAATAGATTGAAAATCGGTTTGCAAGTAGCTGCAGTGAAAGCCCCAGGCTTCGGAGATAACCGCAAGTCCACCCTCAGTGATATGGCTATTGCTACTGGTGGAGTTGTATTTGGTGACGATGCGAATCTCATCAAAATTGAAGATGTCCAACCATCAGACTTAGGTCAAGTAGGTGAAGTGATAATTACTAAAGATGACACTCTGCTATTGAAGGGCAAGGGTAAGAAATCAGACATTGACAGGCGTGCTGAACAGATTAGAGACCAAATTCAGGAGACAAACTCTGAATATGAAAAAGAAAAATTGCAAGAACGTCTGGCTAGACTAGCATCAGGTGTGGCAGTCTTACATGTTGGTGGTTCAAGTGAAGTCGAGGTTAATGAGAAAAAAGACAGAGTTAATGATGCCCTTAATGCTACTAGAGCTGCAGTTGAGGAAGGTATTGTACCTGGAGGTGGTTCTGCACTCTTGAGATGTATTCCAGTATTAGAACAACTCAAAACAGTCAACAGTGATCAGGCCACTGGTGTAGAGATTGTAATGAAAGCTCTGAGGATGCCATGCATGACAATAGCAAAAAATGCAGGCATTGATGGTTCTGTTGTTGTTGCCAAAGTGGAAGACCTTGGAGATGAATTTGGGTATGATGCCCTCAACAATGAATATGTAAACATGATTGAAAAGGGCATCATTGACCCCACAAAGGTGGTGCGGACAGCCCTGACCGATGCCAGTGGTGTTGCATCACTGCTTACTACTGCCGAAGCTGTGATCTGTGAAATTCCACAAGAAAAAGAGCCTAACCCCATGGGTGGTATGGGAGGTATGGGTGGAATGGGTGGTATGGGAGGCATGATGTGA
Protein
MKALMLQGVDILADAVAVTMGPKGRNVILEQSWGSPKITKDGVTVAKGVELKDKFQNIGAKLVQNVANNTNEEAGDGTTTATVLARAIAKEGFEKISKGANPIEIRRGVMLAVDAVKEKLKGMSKPVTTPEEIAQVATISANGDTAIGKLIADAMKKVGRDGVITVKDGKTLTDELEIIEGMKFDRGYISPYFINSSKGAKVEFQDALVLFSEKKISNVQTIIPALEMANQQRKPLIIVAEDVDGEALSTLVVNRLKIGLQVAAVKAPGFGDNRKSTLSDMAIATGGVVFGDDANLIKIEDVQPSDLGQVGEVIITKDDTLLLKGKGKKSDIDRRAEQIRDQIQETNSEYEKEKLQERLARLASGVAVLHVGGSSEVEVNEKKDRVNDALNATRAAVEEGIVPGGGSALLRCIPVLEQLKTVNSDQATGVEIVMKALRMPCMTIAKNAGIDGSVVVAKVEDLGDEFGYDALNNEYVNMIEKGIIDPTKVVRTALTDASGVASLLTTAEAVICEIPQEKEPNPMGGMGGMGGMGGMGGMM
Summary
Description
Prevents misfolding and promotes the refolding and proper assembly of unfolded polypeptides generated under stress conditions.
Similarity
Belongs to the chaperonin (HSP60) family.
Keywords
ATP-binding
Chaperone
Complete proteome
Direct protein sequencing
Mitochondrion
Nucleotide-binding
Reference proteome
Transit peptide
Feature
chain Heat shock protein 60A
Uniprot
H9JCV2
A0A2H4G805
C7ED93
A0A2H1WYZ7
A0A2D1CRQ7
A0A0L7KY35
+ More
A0A194R4W7 A0A194QAN9 A0A2H4LHM4 A0A212FK63 A0A2Z4NA29 A0A1L2EC67 A0A0K0KJ78 A0A336LUG9 O46219 A0A182RBE9 A0A182K8S6 E2APZ5 F4X042 A0A182N6E7 A0A232EQI0 U5EX45 A0A182YID8 A0A195AUN5 A0A158NHK3 E1AC54 A0A182XKJ7 A0A182VA99 A0A195ETM2 A0A182KMN2 A0A182P8X8 A0A182UBV0 A0A2J7QKN9 C1K659 K7J7M0 A0A1J1HTG9 A0A0K8TSS6 Q7Q270 A0A0J7L0Q8 A0A182VXM0 A0A151WGI8 A0A195C3H2 A0A182QQI4 E9J0F3 A0A067QVI3 A0A182F825 A0A2M4A2R6 A0A182JIX3 A0A2M3Z345 A0A2M4BII7 W5J7H7 A0A191UR63 A0A182LRW3 A0A1L8DTR7 E2BX50 A0A026WB63 A0A084W5F2 A0A0K8U4C1 A0A1B6KKQ0 W8CBF7 A0A0K8W6E6 I1SWI8 A0A0A1X785 A0A1I8QA58 A0A240FEJ5 A0A2P8YHT8 A0A1I7PBP8 A0A2A3E2E9 V9II55 A0A088ART2 A0A1A9YJB9 A0A1B0BAY1 A0A171B8K0 A0A1A9URH9 A0A224XHB7 C7LA94 T1IAB6 T1PA52 O02649 A0A1W4V2G3 D3TR05 A0A1L8EH67 A0A069DVR3 A0A346THP7 A0A386GY53 B4R2R2 A0A0V0G3P4 A0A0C9PQ32 A0A1A9W962 A0A023F5L6 A0A1B0A2T9 A0A0A9XLY6 A0A1Q3F3W2 A0A1Q3F3Z0 A0A0L7RDJ2 A0A097PHN0 B3NVA9 B4MAL6 B4JXC6 A0A0B4L061
A0A194R4W7 A0A194QAN9 A0A2H4LHM4 A0A212FK63 A0A2Z4NA29 A0A1L2EC67 A0A0K0KJ78 A0A336LUG9 O46219 A0A182RBE9 A0A182K8S6 E2APZ5 F4X042 A0A182N6E7 A0A232EQI0 U5EX45 A0A182YID8 A0A195AUN5 A0A158NHK3 E1AC54 A0A182XKJ7 A0A182VA99 A0A195ETM2 A0A182KMN2 A0A182P8X8 A0A182UBV0 A0A2J7QKN9 C1K659 K7J7M0 A0A1J1HTG9 A0A0K8TSS6 Q7Q270 A0A0J7L0Q8 A0A182VXM0 A0A151WGI8 A0A195C3H2 A0A182QQI4 E9J0F3 A0A067QVI3 A0A182F825 A0A2M4A2R6 A0A182JIX3 A0A2M3Z345 A0A2M4BII7 W5J7H7 A0A191UR63 A0A182LRW3 A0A1L8DTR7 E2BX50 A0A026WB63 A0A084W5F2 A0A0K8U4C1 A0A1B6KKQ0 W8CBF7 A0A0K8W6E6 I1SWI8 A0A0A1X785 A0A1I8QA58 A0A240FEJ5 A0A2P8YHT8 A0A1I7PBP8 A0A2A3E2E9 V9II55 A0A088ART2 A0A1A9YJB9 A0A1B0BAY1 A0A171B8K0 A0A1A9URH9 A0A224XHB7 C7LA94 T1IAB6 T1PA52 O02649 A0A1W4V2G3 D3TR05 A0A1L8EH67 A0A069DVR3 A0A346THP7 A0A386GY53 B4R2R2 A0A0V0G3P4 A0A0C9PQ32 A0A1A9W962 A0A023F5L6 A0A1B0A2T9 A0A0A9XLY6 A0A1Q3F3W2 A0A1Q3F3Z0 A0A0L7RDJ2 A0A097PHN0 B3NVA9 B4MAL6 B4JXC6 A0A0B4L061
Pubmed
19121390
26227816
26354079
28756777
22118469
11091906
+ More
20798317 21719571 28648823 25244985 21347285 20966253 22517445 20075255 26369729 12364791 14747013 17210077 21282665 24845553 20920257 23761445 27142481 24508170 30249741 24438588 24495485 25830018 28727798 29403074 25315136 10731132 12537572 10071211 8500545 20353571 26334808 30226893 17994087 25474469 25401762 26823975 30659637 18057021
20798317 21719571 28648823 25244985 21347285 20966253 22517445 20075255 26369729 12364791 14747013 17210077 21282665 24845553 20920257 23761445 27142481 24508170 30249741 24438588 24495485 25830018 28727798 29403074 25315136 10731132 12537572 10071211 8500545 20353571 26334808 30226893 17994087 25474469 25401762 26823975 30659637 18057021
EMBL
BABH01027708
KU522481
APX61067.1
GQ265913
ACT52824.1
ODYU01012102
+ More
SOQ58222.1 KX977588 ATN45247.1 JTDY01004454 KOB68143.1 KQ460685 KPJ12848.1 KQ459232 KPJ02499.1 KX845566 KZ150062 ATB54997.1 PZC74202.1 AGBW02008144 OWR54106.1 MG520362 AWX67684.1 KT003962 ANJ86402.1 KM215269 AIJ00866.1 UFQT01000134 SSX20641.1 U87959 AAB94640.1 GL441645 EFN64482.1 GL888489 EGI60183.1 NNAY01002764 OXU20614.1 GANO01001135 JAB58736.1 KQ976737 KYM75953.1 ADTU01015877 HM589532 ADM13383.1 KQ981975 KYN31588.1 NEVH01013262 PNF29143.1 FJ798092 ACO57619.1 AAZX01002179 CVRI01000012 CRK89481.1 GDAI01000420 JAI17183.1 AAAB01008978 EAA13612.2 LBMM01001509 KMQ96173.1 KQ983167 KYQ46956.1 KQ978350 KYM94736.1 AXCN02000286 GL767462 EFZ13709.1 KK852902 KDR14060.1 GGFK01001700 MBW35021.1 GGFM01002162 MBW22913.1 GGFJ01003728 MBW52869.1 ADMH02002112 ETN58825.1 KU365953 ANJ04669.1 AXCM01007531 GFDF01004231 JAV09853.1 GL451202 EFN79769.1 KK107293 QOIP01000005 EZA53300.1 RLU22466.1 ATLV01020615 KE525303 KFB45446.1 GDHF01031164 JAI21150.1 GEBQ01027956 JAT12021.1 GAMC01000549 JAC06007.1 GDHF01005859 JAI46455.1 JF812645 AEG80296.1 GBXI01007128 JAD07164.1 KY231147 ARQ84027.1 PYGN01000583 PSN43817.1 KR733065 AND99759.1 KZ288427 PBC25875.1 JR048942 AEY60808.1 JXJN01011213 GEMB01000139 JAS02973.1 GFTR01007228 JAW09198.1 BT099644 ACV04814.1 ACPB03019478 KA645656 AFP60285.1 X99341 AE014298 BT010206 Y09066 CCAG010015761 EZ423857 ADD20133.1 GFDG01000804 JAV17995.1 GBGD01000993 JAC87896.1 MG265924 AXU24980.1 MH490975 AYD37988.1 CM000366 EDX17611.1 GECL01003438 JAP02686.1 GBYB01003313 JAG73080.1 GBBI01002259 JAC16453.1 GBHO01022618 GBHO01022615 GBRD01012011 GBRD01012009 GDHC01015044 JAG20986.1 JAG20989.1 JAG53813.1 JAQ03585.1 GFDL01012789 JAV22256.1 GFDL01012771 JAV22274.1 KQ414614 KOC68831.1 KM221892 MK139549 AIU47033.1 QAX88055.1 CH954180 EDV46097.1 KQS29825.1 CH940655 EDW66275.1 CH916376 EDV95402.1 KC620435 AHE77384.1
SOQ58222.1 KX977588 ATN45247.1 JTDY01004454 KOB68143.1 KQ460685 KPJ12848.1 KQ459232 KPJ02499.1 KX845566 KZ150062 ATB54997.1 PZC74202.1 AGBW02008144 OWR54106.1 MG520362 AWX67684.1 KT003962 ANJ86402.1 KM215269 AIJ00866.1 UFQT01000134 SSX20641.1 U87959 AAB94640.1 GL441645 EFN64482.1 GL888489 EGI60183.1 NNAY01002764 OXU20614.1 GANO01001135 JAB58736.1 KQ976737 KYM75953.1 ADTU01015877 HM589532 ADM13383.1 KQ981975 KYN31588.1 NEVH01013262 PNF29143.1 FJ798092 ACO57619.1 AAZX01002179 CVRI01000012 CRK89481.1 GDAI01000420 JAI17183.1 AAAB01008978 EAA13612.2 LBMM01001509 KMQ96173.1 KQ983167 KYQ46956.1 KQ978350 KYM94736.1 AXCN02000286 GL767462 EFZ13709.1 KK852902 KDR14060.1 GGFK01001700 MBW35021.1 GGFM01002162 MBW22913.1 GGFJ01003728 MBW52869.1 ADMH02002112 ETN58825.1 KU365953 ANJ04669.1 AXCM01007531 GFDF01004231 JAV09853.1 GL451202 EFN79769.1 KK107293 QOIP01000005 EZA53300.1 RLU22466.1 ATLV01020615 KE525303 KFB45446.1 GDHF01031164 JAI21150.1 GEBQ01027956 JAT12021.1 GAMC01000549 JAC06007.1 GDHF01005859 JAI46455.1 JF812645 AEG80296.1 GBXI01007128 JAD07164.1 KY231147 ARQ84027.1 PYGN01000583 PSN43817.1 KR733065 AND99759.1 KZ288427 PBC25875.1 JR048942 AEY60808.1 JXJN01011213 GEMB01000139 JAS02973.1 GFTR01007228 JAW09198.1 BT099644 ACV04814.1 ACPB03019478 KA645656 AFP60285.1 X99341 AE014298 BT010206 Y09066 CCAG010015761 EZ423857 ADD20133.1 GFDG01000804 JAV17995.1 GBGD01000993 JAC87896.1 MG265924 AXU24980.1 MH490975 AYD37988.1 CM000366 EDX17611.1 GECL01003438 JAP02686.1 GBYB01003313 JAG73080.1 GBBI01002259 JAC16453.1 GBHO01022618 GBHO01022615 GBRD01012011 GBRD01012009 GDHC01015044 JAG20986.1 JAG20989.1 JAG53813.1 JAQ03585.1 GFDL01012789 JAV22256.1 GFDL01012771 JAV22274.1 KQ414614 KOC68831.1 KM221892 MK139549 AIU47033.1 QAX88055.1 CH954180 EDV46097.1 KQS29825.1 CH940655 EDW66275.1 CH916376 EDV95402.1 KC620435 AHE77384.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000075900
+ More
UP000075881 UP000000311 UP000007755 UP000075884 UP000215335 UP000076408 UP000078540 UP000005205 UP000076407 UP000075903 UP000078541 UP000075882 UP000075885 UP000075902 UP000235965 UP000002358 UP000183832 UP000007062 UP000036403 UP000075920 UP000075809 UP000078542 UP000075886 UP000027135 UP000069272 UP000075880 UP000000673 UP000075883 UP000008237 UP000053097 UP000279307 UP000030765 UP000095300 UP000245037 UP000242457 UP000005203 UP000092443 UP000092460 UP000078200 UP000015103 UP000095301 UP000000803 UP000192221 UP000092444 UP000000304 UP000091820 UP000092445 UP000053825 UP000008711 UP000008792 UP000001070
UP000075881 UP000000311 UP000007755 UP000075884 UP000215335 UP000076408 UP000078540 UP000005205 UP000076407 UP000075903 UP000078541 UP000075882 UP000075885 UP000075902 UP000235965 UP000002358 UP000183832 UP000007062 UP000036403 UP000075920 UP000075809 UP000078542 UP000075886 UP000027135 UP000069272 UP000075880 UP000000673 UP000075883 UP000008237 UP000053097 UP000279307 UP000030765 UP000095300 UP000245037 UP000242457 UP000005203 UP000092443 UP000092460 UP000078200 UP000015103 UP000095301 UP000000803 UP000192221 UP000092444 UP000000304 UP000091820 UP000092445 UP000053825 UP000008711 UP000008792 UP000001070
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JCV2
A0A2H4G805
C7ED93
A0A2H1WYZ7
A0A2D1CRQ7
A0A0L7KY35
+ More
A0A194R4W7 A0A194QAN9 A0A2H4LHM4 A0A212FK63 A0A2Z4NA29 A0A1L2EC67 A0A0K0KJ78 A0A336LUG9 O46219 A0A182RBE9 A0A182K8S6 E2APZ5 F4X042 A0A182N6E7 A0A232EQI0 U5EX45 A0A182YID8 A0A195AUN5 A0A158NHK3 E1AC54 A0A182XKJ7 A0A182VA99 A0A195ETM2 A0A182KMN2 A0A182P8X8 A0A182UBV0 A0A2J7QKN9 C1K659 K7J7M0 A0A1J1HTG9 A0A0K8TSS6 Q7Q270 A0A0J7L0Q8 A0A182VXM0 A0A151WGI8 A0A195C3H2 A0A182QQI4 E9J0F3 A0A067QVI3 A0A182F825 A0A2M4A2R6 A0A182JIX3 A0A2M3Z345 A0A2M4BII7 W5J7H7 A0A191UR63 A0A182LRW3 A0A1L8DTR7 E2BX50 A0A026WB63 A0A084W5F2 A0A0K8U4C1 A0A1B6KKQ0 W8CBF7 A0A0K8W6E6 I1SWI8 A0A0A1X785 A0A1I8QA58 A0A240FEJ5 A0A2P8YHT8 A0A1I7PBP8 A0A2A3E2E9 V9II55 A0A088ART2 A0A1A9YJB9 A0A1B0BAY1 A0A171B8K0 A0A1A9URH9 A0A224XHB7 C7LA94 T1IAB6 T1PA52 O02649 A0A1W4V2G3 D3TR05 A0A1L8EH67 A0A069DVR3 A0A346THP7 A0A386GY53 B4R2R2 A0A0V0G3P4 A0A0C9PQ32 A0A1A9W962 A0A023F5L6 A0A1B0A2T9 A0A0A9XLY6 A0A1Q3F3W2 A0A1Q3F3Z0 A0A0L7RDJ2 A0A097PHN0 B3NVA9 B4MAL6 B4JXC6 A0A0B4L061
A0A194R4W7 A0A194QAN9 A0A2H4LHM4 A0A212FK63 A0A2Z4NA29 A0A1L2EC67 A0A0K0KJ78 A0A336LUG9 O46219 A0A182RBE9 A0A182K8S6 E2APZ5 F4X042 A0A182N6E7 A0A232EQI0 U5EX45 A0A182YID8 A0A195AUN5 A0A158NHK3 E1AC54 A0A182XKJ7 A0A182VA99 A0A195ETM2 A0A182KMN2 A0A182P8X8 A0A182UBV0 A0A2J7QKN9 C1K659 K7J7M0 A0A1J1HTG9 A0A0K8TSS6 Q7Q270 A0A0J7L0Q8 A0A182VXM0 A0A151WGI8 A0A195C3H2 A0A182QQI4 E9J0F3 A0A067QVI3 A0A182F825 A0A2M4A2R6 A0A182JIX3 A0A2M3Z345 A0A2M4BII7 W5J7H7 A0A191UR63 A0A182LRW3 A0A1L8DTR7 E2BX50 A0A026WB63 A0A084W5F2 A0A0K8U4C1 A0A1B6KKQ0 W8CBF7 A0A0K8W6E6 I1SWI8 A0A0A1X785 A0A1I8QA58 A0A240FEJ5 A0A2P8YHT8 A0A1I7PBP8 A0A2A3E2E9 V9II55 A0A088ART2 A0A1A9YJB9 A0A1B0BAY1 A0A171B8K0 A0A1A9URH9 A0A224XHB7 C7LA94 T1IAB6 T1PA52 O02649 A0A1W4V2G3 D3TR05 A0A1L8EH67 A0A069DVR3 A0A346THP7 A0A386GY53 B4R2R2 A0A0V0G3P4 A0A0C9PQ32 A0A1A9W962 A0A023F5L6 A0A1B0A2T9 A0A0A9XLY6 A0A1Q3F3W2 A0A1Q3F3Z0 A0A0L7RDJ2 A0A097PHN0 B3NVA9 B4MAL6 B4JXC6 A0A0B4L061
PDB
4PJ1
E-value=0,
Score=1772
Ontologies
GO
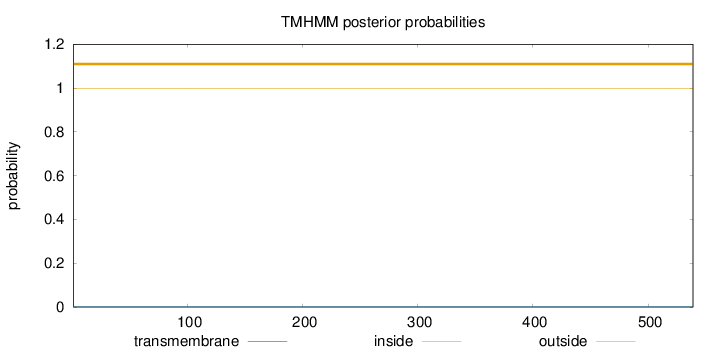
Topology
Subcellular location
Mitochondrion matrix
Length:
539
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00386
Exp number, first 60 AAs:
0.00116
Total prob of N-in:
0.00103
outside
1 - 539
Population Genetic Test Statistics
Pi
165.835708
Theta
192.973714
Tajima's D
-0.968671
CLR
39.056498
CSRT
0.144692765361732
Interpretation
Uncertain
