Pre Gene Modal
BGIBMGA007352
Annotation
PREDICTED:_uncharacterized_protein_LOC106116963_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.389 Extracellular Reliability : 1.395
Sequence
CDS
ATGAAAAAGCGTTTCATGCGGAAGCCAAATGTTTCTGAAGCAACAAATGAGTTTATAGCACTAGCGATACAATGTGAACATTCTGAACAGCCAACGTTTGCAGGGCATTCTTATGTGGGCGCTGCGAAATGCGAGGCCTCGGTGGGAAATTTCCTTGGTGAAGCAGAGCACTATATCTCTGCAGCCAGACAATTCATTAAAGCTGAGATGAAACTGAAGTCAATGAAATTCTACAGTCCCGAAAGAGAGAATTTAGAAGCTGGTATAGGATGTTTTCTACAAGCACTGAACAAATATCCAGAAAAGTCACCCATTCGCACATCACTACTGATGGAATTAGCTAGCAACCTGTCCACCCTGGGACATAAAGCGGAGGCTATATCATACTACCAACAAGCACTGGATACTGTTGTGGATTATACTATGAAGTTGATGGCATTGTGGAATTTAATGATTTTACAGATTGACAGTGAAAAGTTCAGCATGGCACTTGAAACTTCAAACACAATATGTGACTCCAAACTGAATATTCCTGAAGACCTCCTCACAGAAGTTCAAGTAACCAGGATACTGTTAGCAATTCTCGTGAAACCAGCCGATGAGGACAAGCCATCTTCATTGAAACAAGTATTTTTTGATCTAATGAATGATATAGAAACTGAAGCTCTACCTCTGAGTACGGACCTGCGGTTGAAACTGCAGTCAATAACAGTGAGCGAGCAGGAGGGAGATATGACGTCATTGGTTGCACTATTGGCCGACACGGAAGAGCACCTAGTTCCACACCAGACACAATTACTGCACCACATTATTTCTAAACAAAGGCTCGATCATTTGGGTCTTACATAA
Protein
MKKRFMRKPNVSEATNEFIALAIQCEHSEQPTFAGHSYVGAAKCEASVGNFLGEAEHYISAARQFIKAEMKLKSMKFYSPERENLEAGIGCFLQALNKYPEKSPIRTSLLMELASNLSTLGHKAEAISYYQQALDTVVDYTMKLMALWNLMILQIDSEKFSMALETSNTICDSKLNIPEDLLTEVQVTRILLAILVKPADEDKPSSLKQVFFDLMNDIETEALPLSTDLRLKLQSITVSEQEGDMTSLVALLADTEEHLVPHQTQLLHHIISKQRLDHLGLT
Summary
Cofactor
NAD(+)
Similarity
Belongs to the NAD(P)-dependent epimerase/dehydratase family.
Uniprot
H9JCV5
A0A2A4J1A6
A0A2W1BMV2
A0A194QBX8
A0A194R5Q4
I4DK93
+ More
S4Q089 A0A212FK48 A0A0L7KNJ5 A0A067R0E6 A0A139WE85 A0A1Y1M4Y0 A0A2P8Y0M4 A0A0T6AVQ3 A0A2T7Q1I9 R7TF82 A0A1Y1LZX9 A0A210PFF1 A0A1B6DSH5 A0A1W4X258 V3ZNZ8 A0A2J7RL09 A0A1S3IQY1 C3XYY5 A0A2C9LWC8 A0A1B6HM47 A0A0K2TGW3 E2C0D0 A0A1B6E0R3 J9LB20 A0A154PS13 A0A369SAL5 A0A2H8TH78 A0A0L8IF76 A0A0L8IF40 E9IJP2 A0A2S2NZT2 A0A1B6M889 A0A195FMZ5 J3JX32 K7JCU8 T1I7A1 A0A158NH10 A0A2B4RRW2 A0A023GI10 A0A0C9R2D2 A0A2G8KN29 G3MNG0 A0A3M6UAF0 A0A224YDZ1 E2ARZ8 A0A3N0YKL3 A0A088AHU5 A2BIC9 A0A195D1D1 A0A224YF30 A0A131YT70 A0A2S2QQU2 A0A026WYW6 F4WSS9 T2MG25 B3RN17 A0A131XJS2 A0A2A3EBC9 A0A0J7L496 A0A195B7C5 A7RY05 N6TC88 U4UQN4 A0A0A9WIH7 S4RW38 W4ZIX0 A0A293L5V6 Q0P4L3 Q5BKG4 A0JMS8 K1QXM0 A0A2D0Q2Y5 A0A2R5LL10 G3PTZ7 A0A1L8GNE4 A0A3B4EBD6 M4ASW5 H3AAP2 T1JBR8 W5JJI9 A0A146VEY3 A0A182JZK2 A0A1A7W9K4 A0A3B3WKX5 I3KNJ0
S4Q089 A0A212FK48 A0A0L7KNJ5 A0A067R0E6 A0A139WE85 A0A1Y1M4Y0 A0A2P8Y0M4 A0A0T6AVQ3 A0A2T7Q1I9 R7TF82 A0A1Y1LZX9 A0A210PFF1 A0A1B6DSH5 A0A1W4X258 V3ZNZ8 A0A2J7RL09 A0A1S3IQY1 C3XYY5 A0A2C9LWC8 A0A1B6HM47 A0A0K2TGW3 E2C0D0 A0A1B6E0R3 J9LB20 A0A154PS13 A0A369SAL5 A0A2H8TH78 A0A0L8IF76 A0A0L8IF40 E9IJP2 A0A2S2NZT2 A0A1B6M889 A0A195FMZ5 J3JX32 K7JCU8 T1I7A1 A0A158NH10 A0A2B4RRW2 A0A023GI10 A0A0C9R2D2 A0A2G8KN29 G3MNG0 A0A3M6UAF0 A0A224YDZ1 E2ARZ8 A0A3N0YKL3 A0A088AHU5 A2BIC9 A0A195D1D1 A0A224YF30 A0A131YT70 A0A2S2QQU2 A0A026WYW6 F4WSS9 T2MG25 B3RN17 A0A131XJS2 A0A2A3EBC9 A0A0J7L496 A0A195B7C5 A7RY05 N6TC88 U4UQN4 A0A0A9WIH7 S4RW38 W4ZIX0 A0A293L5V6 Q0P4L3 Q5BKG4 A0JMS8 K1QXM0 A0A2D0Q2Y5 A0A2R5LL10 G3PTZ7 A0A1L8GNE4 A0A3B4EBD6 M4ASW5 H3AAP2 T1JBR8 W5JJI9 A0A146VEY3 A0A182JZK2 A0A1A7W9K4 A0A3B3WKX5 I3KNJ0
Pubmed
19121390
28756777
26354079
22651552
23622113
22118469
+ More
26227816 24845553 18362917 19820115 28004739 29403074 23254933 28812685 18563158 15562597 20798317 30042472 21282665 22516182 20075255 21347285 26131772 29023486 22216098 30382153 28797301 23594743 26830274 24508170 21719571 24065732 18719581 28049606 17615350 23537049 25401762 26823975 22992520 27762356 23542700 9215903 20920257 23761445 25186727
26227816 24845553 18362917 19820115 28004739 29403074 23254933 28812685 18563158 15562597 20798317 30042472 21282665 22516182 20075255 21347285 26131772 29023486 22216098 30382153 28797301 23594743 26830274 24508170 21719571 24065732 18719581 28049606 17615350 23537049 25401762 26823975 22992520 27762356 23542700 9215903 20920257 23761445 25186727
EMBL
BABH01027703
NWSH01004393
PCG65163.1
KZ150062
PZC74206.1
KQ459232
+ More
KPJ02495.1 KQ460685 KPJ12844.1 AK401711 BAM18333.1 GAIX01000208 JAA92352.1 AGBW02008144 OWR54102.1 JTDY01007890 KOB64878.1 KK852804 KDR16201.1 KQ971357 KYB26177.1 GEZM01044365 JAV78277.1 PYGN01001081 PSN37779.1 LJIG01022746 KRT78933.1 PZQS01000001 PVD39529.1 AMQN01002874 KB310235 ELT92152.1 GEZM01044364 JAV78278.1 NEDP02076736 OWF35223.1 GEDC01008661 JAS28637.1 KB203566 ESO84215.1 NEVH01002701 PNF41526.1 GG666474 EEN66756.1 GECU01031943 JAS75763.1 HACA01007370 CDW24731.1 GL451770 EFN78589.1 GEDC01005790 JAS31508.1 ABLF02034920 KQ435119 KZC14715.1 NOWV01000032 RDD43880.1 GFXV01001586 MBW13391.1 KQ415849 KOG00108.1 KOG00106.1 GL763877 EFZ19214.1 GGMR01010100 MBY22719.1 GEBQ01007856 JAT32121.1 KQ981387 KYN41975.1 BT127800 AEE62762.1 AAZX01003993 ACPB03002264 ADTU01015424 LSMT01000382 PFX18972.1 GBBM01001877 JAC33541.1 GBZX01001555 JAG91185.1 MRZV01000466 PIK49424.1 JO843411 AEO35028.1 RCHS01001930 RMX50643.1 GFPF01001267 MAA12413.1 GL442209 EFN63772.1 RJVU01037554 ROL46450.1 BX901927 CU570697 KQ976973 KYN06656.1 GFPF01001266 MAA12412.1 GEDV01006144 JAP82413.1 GGMS01010902 MBY80105.1 KK107063 EZA61217.1 GL888327 EGI62736.1 HAAD01004981 CDG71213.1 DS985242 EDV27377.1 GEFH01002800 JAP65781.1 KZ288310 PBC28602.1 LBMM01000855 KMQ97363.1 KQ976574 KYM80099.1 DS469552 EDO43613.1 APGK01035707 KB740928 ENN77919.1 KB632428 ERL95412.1 GBHO01037276 GBRD01011757 GDHC01013082 JAG06328.1 JAG54067.1 JAQ05547.1 AAGJ04057522 AAGJ04057523 AAGJ04057524 AAGJ04057525 GFWV01002387 MAA27117.1 BC122015 AAI22016.1 BC091084 AAH91084.1 BC125991 AAI25992.1 JH818186 EKC41637.1 GGLE01006055 MBY10181.1 CM004472 OCT85339.1 AFYH01234871 AFYH01234872 AFYH01234873 AFYH01234874 AFYH01234875 JH432018 ADMH02001051 ETN64306.1 GCES01070360 JAR15963.1 HADW01000804 HADX01009604 SBP02204.1 AERX01018827
KPJ02495.1 KQ460685 KPJ12844.1 AK401711 BAM18333.1 GAIX01000208 JAA92352.1 AGBW02008144 OWR54102.1 JTDY01007890 KOB64878.1 KK852804 KDR16201.1 KQ971357 KYB26177.1 GEZM01044365 JAV78277.1 PYGN01001081 PSN37779.1 LJIG01022746 KRT78933.1 PZQS01000001 PVD39529.1 AMQN01002874 KB310235 ELT92152.1 GEZM01044364 JAV78278.1 NEDP02076736 OWF35223.1 GEDC01008661 JAS28637.1 KB203566 ESO84215.1 NEVH01002701 PNF41526.1 GG666474 EEN66756.1 GECU01031943 JAS75763.1 HACA01007370 CDW24731.1 GL451770 EFN78589.1 GEDC01005790 JAS31508.1 ABLF02034920 KQ435119 KZC14715.1 NOWV01000032 RDD43880.1 GFXV01001586 MBW13391.1 KQ415849 KOG00108.1 KOG00106.1 GL763877 EFZ19214.1 GGMR01010100 MBY22719.1 GEBQ01007856 JAT32121.1 KQ981387 KYN41975.1 BT127800 AEE62762.1 AAZX01003993 ACPB03002264 ADTU01015424 LSMT01000382 PFX18972.1 GBBM01001877 JAC33541.1 GBZX01001555 JAG91185.1 MRZV01000466 PIK49424.1 JO843411 AEO35028.1 RCHS01001930 RMX50643.1 GFPF01001267 MAA12413.1 GL442209 EFN63772.1 RJVU01037554 ROL46450.1 BX901927 CU570697 KQ976973 KYN06656.1 GFPF01001266 MAA12412.1 GEDV01006144 JAP82413.1 GGMS01010902 MBY80105.1 KK107063 EZA61217.1 GL888327 EGI62736.1 HAAD01004981 CDG71213.1 DS985242 EDV27377.1 GEFH01002800 JAP65781.1 KZ288310 PBC28602.1 LBMM01000855 KMQ97363.1 KQ976574 KYM80099.1 DS469552 EDO43613.1 APGK01035707 KB740928 ENN77919.1 KB632428 ERL95412.1 GBHO01037276 GBRD01011757 GDHC01013082 JAG06328.1 JAG54067.1 JAQ05547.1 AAGJ04057522 AAGJ04057523 AAGJ04057524 AAGJ04057525 GFWV01002387 MAA27117.1 BC122015 AAI22016.1 BC091084 AAH91084.1 BC125991 AAI25992.1 JH818186 EKC41637.1 GGLE01006055 MBY10181.1 CM004472 OCT85339.1 AFYH01234871 AFYH01234872 AFYH01234873 AFYH01234874 AFYH01234875 JH432018 ADMH02001051 ETN64306.1 GCES01070360 JAR15963.1 HADW01000804 HADX01009604 SBP02204.1 AERX01018827
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000027135 UP000007266 UP000245037 UP000245119 UP000014760 UP000242188 UP000192223 UP000030746 UP000235965 UP000085678 UP000001554 UP000076420 UP000008237 UP000007819 UP000076502 UP000253843 UP000053454 UP000078541 UP000002358 UP000015103 UP000005205 UP000225706 UP000230750 UP000275408 UP000000311 UP000005203 UP000000437 UP000078542 UP000053097 UP000007755 UP000009022 UP000242457 UP000036403 UP000078540 UP000001593 UP000019118 UP000030742 UP000245300 UP000007110 UP000005408 UP000221080 UP000007635 UP000186698 UP000261440 UP000002852 UP000008672 UP000000673 UP000075881 UP000261480 UP000005207
UP000027135 UP000007266 UP000245037 UP000245119 UP000014760 UP000242188 UP000192223 UP000030746 UP000235965 UP000085678 UP000001554 UP000076420 UP000008237 UP000007819 UP000076502 UP000253843 UP000053454 UP000078541 UP000002358 UP000015103 UP000005205 UP000225706 UP000230750 UP000275408 UP000000311 UP000005203 UP000000437 UP000078542 UP000053097 UP000007755 UP000009022 UP000242457 UP000036403 UP000078540 UP000001593 UP000019118 UP000030742 UP000245300 UP000007110 UP000005408 UP000221080 UP000007635 UP000186698 UP000261440 UP000002852 UP000008672 UP000000673 UP000075881 UP000261480 UP000005207
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JCV5
A0A2A4J1A6
A0A2W1BMV2
A0A194QBX8
A0A194R5Q4
I4DK93
+ More
S4Q089 A0A212FK48 A0A0L7KNJ5 A0A067R0E6 A0A139WE85 A0A1Y1M4Y0 A0A2P8Y0M4 A0A0T6AVQ3 A0A2T7Q1I9 R7TF82 A0A1Y1LZX9 A0A210PFF1 A0A1B6DSH5 A0A1W4X258 V3ZNZ8 A0A2J7RL09 A0A1S3IQY1 C3XYY5 A0A2C9LWC8 A0A1B6HM47 A0A0K2TGW3 E2C0D0 A0A1B6E0R3 J9LB20 A0A154PS13 A0A369SAL5 A0A2H8TH78 A0A0L8IF76 A0A0L8IF40 E9IJP2 A0A2S2NZT2 A0A1B6M889 A0A195FMZ5 J3JX32 K7JCU8 T1I7A1 A0A158NH10 A0A2B4RRW2 A0A023GI10 A0A0C9R2D2 A0A2G8KN29 G3MNG0 A0A3M6UAF0 A0A224YDZ1 E2ARZ8 A0A3N0YKL3 A0A088AHU5 A2BIC9 A0A195D1D1 A0A224YF30 A0A131YT70 A0A2S2QQU2 A0A026WYW6 F4WSS9 T2MG25 B3RN17 A0A131XJS2 A0A2A3EBC9 A0A0J7L496 A0A195B7C5 A7RY05 N6TC88 U4UQN4 A0A0A9WIH7 S4RW38 W4ZIX0 A0A293L5V6 Q0P4L3 Q5BKG4 A0JMS8 K1QXM0 A0A2D0Q2Y5 A0A2R5LL10 G3PTZ7 A0A1L8GNE4 A0A3B4EBD6 M4ASW5 H3AAP2 T1JBR8 W5JJI9 A0A146VEY3 A0A182JZK2 A0A1A7W9K4 A0A3B3WKX5 I3KNJ0
S4Q089 A0A212FK48 A0A0L7KNJ5 A0A067R0E6 A0A139WE85 A0A1Y1M4Y0 A0A2P8Y0M4 A0A0T6AVQ3 A0A2T7Q1I9 R7TF82 A0A1Y1LZX9 A0A210PFF1 A0A1B6DSH5 A0A1W4X258 V3ZNZ8 A0A2J7RL09 A0A1S3IQY1 C3XYY5 A0A2C9LWC8 A0A1B6HM47 A0A0K2TGW3 E2C0D0 A0A1B6E0R3 J9LB20 A0A154PS13 A0A369SAL5 A0A2H8TH78 A0A0L8IF76 A0A0L8IF40 E9IJP2 A0A2S2NZT2 A0A1B6M889 A0A195FMZ5 J3JX32 K7JCU8 T1I7A1 A0A158NH10 A0A2B4RRW2 A0A023GI10 A0A0C9R2D2 A0A2G8KN29 G3MNG0 A0A3M6UAF0 A0A224YDZ1 E2ARZ8 A0A3N0YKL3 A0A088AHU5 A2BIC9 A0A195D1D1 A0A224YF30 A0A131YT70 A0A2S2QQU2 A0A026WYW6 F4WSS9 T2MG25 B3RN17 A0A131XJS2 A0A2A3EBC9 A0A0J7L496 A0A195B7C5 A7RY05 N6TC88 U4UQN4 A0A0A9WIH7 S4RW38 W4ZIX0 A0A293L5V6 Q0P4L3 Q5BKG4 A0JMS8 K1QXM0 A0A2D0Q2Y5 A0A2R5LL10 G3PTZ7 A0A1L8GNE4 A0A3B4EBD6 M4ASW5 H3AAP2 T1JBR8 W5JJI9 A0A146VEY3 A0A182JZK2 A0A1A7W9K4 A0A3B3WKX5 I3KNJ0
PDB
6EZ8
E-value=7.73009e-05,
Score=108
Ontologies
GO
PANTHER
Topology
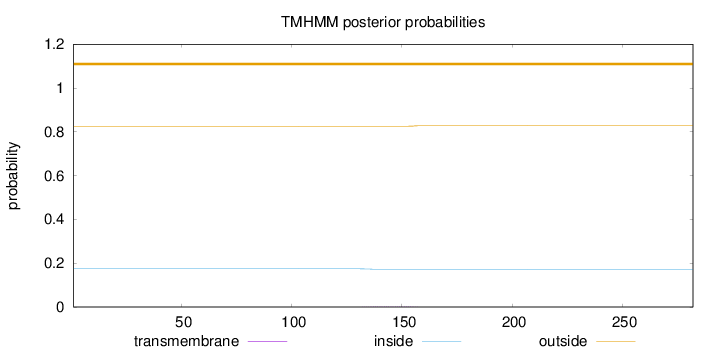
Length:
282
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04676
Exp number, first 60 AAs:
0.00078
Total prob of N-in:
0.17449
outside
1 - 282
Population Genetic Test Statistics
Pi
206.088312
Theta
180.983198
Tajima's D
1.251832
CLR
0.2179
CSRT
0.724363781810909
Interpretation
Uncertain
