Pre Gene Modal
BGIBMGA007413
Annotation
PREDICTED:_serine/threonine-protein_kinase_RIO3_[Papilio_machaon]
Full name
Serine/threonine-protein kinase RIO3
+ More
Inhibitor of growth protein
Inhibitor of growth protein
Location in the cell
Nuclear Reliability : 4.228
Sequence
CDS
ATGTCGAATCCTTGGAAAAAAGTGACAGCACCAGTAGAGGCGCAGAAGCTGTCGGATATCATGTCTGAAGAATTTGCAAGAGACCTTCAGGTTAGAGAAGAGATAAAATTTGCAGAACATCTCTCTGAGCACCATGTCGGCACCTCTAATGAAGTTCCAGTTGAGTTGTTACAAGAAATAGAAAGCTGCAACGCAAAGGAATATTGTGACTCTGACGCGATTATTGCAAAAGTTTTGCAATGTCAGTTCGACAGAGAACACGATGACGAAATCAAACGCGTAGAGAAAAAACGTAATGGAGATGCTAAAGTATCCGTTTCCTATGATAATTACCGCAACGTACCATCGCATTTGATTTACGACTCTGAAGACGACGATGACTTTGAGGGCGATAAGAAAGACTGGGATCGCTTCGAAACAAACGAAAAGGAATTCGCAAGCCTTCCGAAACGCGGATTTATCATAAAAGATGGAGCAATGGTCACCAAACACGACAGTGTTATCACTGGAAGGCGGAATGCTTGTAAAGTTATGGCATTCCCTCCGGAAGTATGCACTGGAGATGGCGCTGGCTTCGATATGAAACTCTCCAATTCTGTATTTAATCATTTGAAAGAACAGACGAGATATCACAATACACGTAAACATAAAATGTTGGACAGAAAGGAGAGTCAAGCTACAGCCGAAATGGGCCTGGATGAGCCTACTAGATTGATACTATTCAAACTTATTAACAATGGGGTATTAGAGGATATCAATGGTGTCATCTCAACAGGAAAGGAGTCTGTTGTTCTACATGCTAATGGTGACACAACTTATCCTGACTTAGATGTACCTAGAGAATGTGCTATTAAAGTCTTCAAAACAACTCTTAACGAGTTCAAAACTCGAGACAAATACATAGAAGCCGATTATAGATTCAAAGATCGCTTCTCGAAACAAAATCCAAGAAAAATTGTACACATGTGGGCTGAGAAGGAGATGCACAACATGATGCGCTTGCAGAAGATAGGATTGAACTGTCCGGATATGATCTGCTTGAAGAAACATATACTTTTAATGTCATTTATTGGGAAGGATAACAAGCCAGCCCCCAAGCTAAGAGACATAATCTTAAGGTCAGATAAATGGCACAGTGTTTACAATGAGGTTGTGGAGAGTATGCATAAAATGTACAATGTTGGTCATCTGATTCATGCTGATCTATCAGAGTATAACATTCTCTGGTGGGATAATAAATGCTGGTTTATTGATGTTTCTCAGTCTGTGCAACCTGACCACCCTAACGGGTTGGAGTTTTTACTTCGAGATTGTCGTAATATTGTCAATTTCTTTGAAAAAAAGGGGGTCTCACATGTGCAAACTGCAGAAGAACTTTTTAAATCAATAACCGGCTTTAATGAAGTTGATGTGAACTTATTGGAAGGAGTTCATACCACATACAATTCTCTGGCATGCCGATTTGAAATCGCCCCCGATGATAATCGAAACGTTAGCTATCCATTTGAATATTGTTGGCAGAAAACCAATGAAGAGAAAGCTAAAAGTGATAAAAGTGATGATAATGAAGAAGATGAATTTGAGGAAATGTGGAATCATGCTGCCAAAAGTAAAACTGTTGATAGTGCTGCCAATTTTGGTAATGAAGTCAAGATTGACAATGAAATAACTTCAACAGCTTCTAAAACCTCTGATTGTAGAGATATTATCGATGATGAATCATGTTTTGGTAATGAATTTAAGGGTACTATACCCACAAATTCAGACGGAGACACCTCTAGTAGTAGAGTAGAGAAAAAGTCTTAA
Protein
MSNPWKKVTAPVEAQKLSDIMSEEFARDLQVREEIKFAEHLSEHHVGTSNEVPVELLQEIESCNAKEYCDSDAIIAKVLQCQFDREHDDEIKRVEKKRNGDAKVSVSYDNYRNVPSHLIYDSEDDDDFEGDKKDWDRFETNEKEFASLPKRGFIIKDGAMVTKHDSVITGRRNACKVMAFPPEVCTGDGAGFDMKLSNSVFNHLKEQTRYHNTRKHKMLDRKESQATAEMGLDEPTRLILFKLINNGVLEDINGVISTGKESVVLHANGDTTYPDLDVPRECAIKVFKTTLNEFKTRDKYIEADYRFKDRFSKQNPRKIVHMWAEKEMHNMMRLQKIGLNCPDMICLKKHILLMSFIGKDNKPAPKLRDIILRSDKWHSVYNEVVESMHKMYNVGHLIHADLSEYNILWWDNKCWFIDVSQSVQPDHPNGLEFLLRDCRNIVNFFEKKGVSHVQTAEELFKSITGFNEVDVNLLEGVHTTYNSLACRFEIAPDDNRNVSYPFEYCWQKTNEEKAKSDKSDDNEEDEFEEMWNHAAKSKTVDSAANFGNEVKIDNEITSTASKTSDCRDIIDDESCFGNEFKGTIPTNSDGDTSSSRVEKKS
Summary
Catalytic Activity
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Subunit
Interacts with H3K4me3 and to a lesser extent with H3K4me2.
Similarity
Belongs to the protein kinase superfamily. RIO-type Ser/Thr kinase family.
Belongs to the ING family.
Belongs to the ING family.
Uniprot
H9JD16
A0A194R4W0
A0A2W1BLP5
A0A194QAN4
A0A2A4J0W2
A0A2H1WAA3
+ More
A0A1E1WK44 A0A0L7LQ88 S4PMA0 A0A212FK40 A0A0K8TM27 A0A1L8DLT7 A0A034V663 A0A067QUR6 A0A0K8UMG9 A0A2P8XG71 Q7Q135 A0A2J7Q9X8 A0A182P7Z4 A0A182K953 W8C3V4 A0A182UV86 Q16VU9 A0A182W8R1 A0A1Q3G358 A0A1S4FMF7 A0A182TT67 A0A182KZ44 A0A182GLE5 A0A182XGY5 A0A182Y699 A0A182G6J3 A0A0A1X499 A0A182RWQ7 A0A182LXC9 B4N0J4 A0A1B0D5R0 A0A1S4J7U9 A0A182F554 A0A1Y1LDC5 A0A084VSR4 A0A182HR90 T1PAS4 W5J4J9 A0A2M4AHY0 A0A1I8ML05 A0A2M4BHW8 A0A2M4BI49 A0A2M4AIH5 A0A182Q2S1 A0A2M4AJF5 T1E355 Q29M98 B4G9J4 A0A182NP21 B3MVJ5 A0A0L0CQJ0 A0A0J9QVS9 A0A1W4UIE4 B4NZ24 B4I178 B4MDQ5 A0A0M4E7S5 Q9VR42 A0A1A9UR86 A0A2M4BJ61 B3N4B2 A0A1B0CT99 A0A182IVQ1 A0A1A9YPR6 A0A1I8P092 A0A1B0BFF2 A0A1A9WUJ8 A0A3B0K0K4 B0W708 A0A1A9Z542 D6WVA9 A0A0T6AT21 B4KGQ8 B4JQA5 A0A1B0FN97 E9GFI5 U5EX34 A0A0P5YUC9 A0A1W4X9G6 A0A0P6ESE4 A0A1D2MN83 A0A336JWB3 A0A0P6BNV1 A0A0P5VJF9 A0A218V1A6 A0A3M0KS59 A0A091KSY3 A0A093FYQ7 U3JUF5 A0A2I0MX49 A0A091JRL0 A0A1V4KFX1 K7FFS5 A0A2I0MX50
A0A1E1WK44 A0A0L7LQ88 S4PMA0 A0A212FK40 A0A0K8TM27 A0A1L8DLT7 A0A034V663 A0A067QUR6 A0A0K8UMG9 A0A2P8XG71 Q7Q135 A0A2J7Q9X8 A0A182P7Z4 A0A182K953 W8C3V4 A0A182UV86 Q16VU9 A0A182W8R1 A0A1Q3G358 A0A1S4FMF7 A0A182TT67 A0A182KZ44 A0A182GLE5 A0A182XGY5 A0A182Y699 A0A182G6J3 A0A0A1X499 A0A182RWQ7 A0A182LXC9 B4N0J4 A0A1B0D5R0 A0A1S4J7U9 A0A182F554 A0A1Y1LDC5 A0A084VSR4 A0A182HR90 T1PAS4 W5J4J9 A0A2M4AHY0 A0A1I8ML05 A0A2M4BHW8 A0A2M4BI49 A0A2M4AIH5 A0A182Q2S1 A0A2M4AJF5 T1E355 Q29M98 B4G9J4 A0A182NP21 B3MVJ5 A0A0L0CQJ0 A0A0J9QVS9 A0A1W4UIE4 B4NZ24 B4I178 B4MDQ5 A0A0M4E7S5 Q9VR42 A0A1A9UR86 A0A2M4BJ61 B3N4B2 A0A1B0CT99 A0A182IVQ1 A0A1A9YPR6 A0A1I8P092 A0A1B0BFF2 A0A1A9WUJ8 A0A3B0K0K4 B0W708 A0A1A9Z542 D6WVA9 A0A0T6AT21 B4KGQ8 B4JQA5 A0A1B0FN97 E9GFI5 U5EX34 A0A0P5YUC9 A0A1W4X9G6 A0A0P6ESE4 A0A1D2MN83 A0A336JWB3 A0A0P6BNV1 A0A0P5VJF9 A0A218V1A6 A0A3M0KS59 A0A091KSY3 A0A093FYQ7 U3JUF5 A0A2I0MX49 A0A091JRL0 A0A1V4KFX1 K7FFS5 A0A2I0MX50
EC Number
2.7.11.1
Pubmed
19121390
26354079
28756777
26227816
23622113
22118469
+ More
26369729 25348373 24845553 29403074 12364791 24495485 17510324 20966253 26483478 25244985 25830018 17994087 28004739 24438588 20920257 23761445 25315136 24330624 15632085 18057021 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 21292972 27289101 23371554 17381049
26369729 25348373 24845553 29403074 12364791 24495485 17510324 20966253 26483478 25244985 25830018 17994087 28004739 24438588 20920257 23761445 25315136 24330624 15632085 18057021 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 21292972 27289101 23371554 17381049
EMBL
BABH01027703
KQ460685
KPJ12843.1
KZ150062
PZC74207.1
KQ459232
+ More
KPJ02494.1 NWSH01004393 PCG65164.1 ODYU01007262 SOQ49886.1 GDQN01003823 JAT87231.1 JTDY01000344 KOB77610.1 GAIX01003680 JAA88880.1 AGBW02008144 OWR54101.1 GDAI01002422 JAI15181.1 GFDF01006672 JAV07412.1 GAKP01021899 JAC37053.1 KK852910 KDR13927.1 GDHF01024749 JAI27565.1 PYGN01002247 PSN31013.1 AAAB01008980 EAA13880.4 NEVH01016341 PNF25400.1 GAMC01005885 GAMC01005884 GAMC01005881 GAMC01005879 JAC00672.1 CH477580 EAT38710.1 GFDL01000803 JAV34242.1 JXUM01071880 KQ562689 KXJ75327.1 JXUM01007600 KQ560243 KXJ83629.1 GBXI01012959 GBXI01008707 JAD01333.1 JAD05585.1 AXCM01000709 CH963920 EDW77607.1 AJVK01025533 GEZM01058838 JAV71652.1 ATLV01016137 KE525057 KFB41008.1 APCN01001659 KA645018 AFP59647.1 ADMH02002089 ETN59392.1 GGFK01007075 MBW40396.1 GGFJ01003491 MBW52632.1 GGFJ01003490 MBW52631.1 GGFK01007087 MBW40408.1 AXCN02001964 GGFK01007599 MBW40920.1 GALA01000134 JAA94718.1 CH379060 EAL33796.2 CH479180 EDW29024.1 CH902624 EDV33260.1 KPU74376.1 JRES01000044 KNC34640.1 CM002910 KMY88157.1 CM000157 EDW87688.1 CH480820 EDW54285.1 CH940661 EDW71316.1 CP012523 ALC37938.1 ALC38425.1 AE014134 AY071312 AAF50965.1 AAL48934.1 GGFJ01003903 MBW53044.1 CH954177 EDV57782.1 AJWK01027315 JXJN01013398 OUUW01000004 SPP79146.1 DS231851 EDS37282.1 KQ971363 EFA07752.1 LJIG01022899 KRT78166.1 CH933807 EDW13258.1 CH916372 EDV99085.1 CCAG010010817 GL732542 EFX81645.1 GANO01000262 JAB59609.1 GDIP01054565 LRGB01001161 JAM49150.1 KZS13409.1 GDIQ01059064 JAN35673.1 LJIJ01000809 ODM94398.1 UFQS01000003 UFQT01000003 SSW96759.1 SSX17146.1 GDIP01012440 JAM91275.1 GDIP01099149 JAM04566.1 MUZQ01000072 OWK59716.1 QRBI01000104 RMC15511.1 KK755440 KFP43112.1 KL214831 KFV61958.1 AGTO01010370 AKCR02000001 PKK34258.1 KK502398 KFP23242.1 LSYS01003169 OPJ83370.1 AGCU01046922 AGCU01046923 AGCU01046924 AGCU01046925 PKK34257.1
KPJ02494.1 NWSH01004393 PCG65164.1 ODYU01007262 SOQ49886.1 GDQN01003823 JAT87231.1 JTDY01000344 KOB77610.1 GAIX01003680 JAA88880.1 AGBW02008144 OWR54101.1 GDAI01002422 JAI15181.1 GFDF01006672 JAV07412.1 GAKP01021899 JAC37053.1 KK852910 KDR13927.1 GDHF01024749 JAI27565.1 PYGN01002247 PSN31013.1 AAAB01008980 EAA13880.4 NEVH01016341 PNF25400.1 GAMC01005885 GAMC01005884 GAMC01005881 GAMC01005879 JAC00672.1 CH477580 EAT38710.1 GFDL01000803 JAV34242.1 JXUM01071880 KQ562689 KXJ75327.1 JXUM01007600 KQ560243 KXJ83629.1 GBXI01012959 GBXI01008707 JAD01333.1 JAD05585.1 AXCM01000709 CH963920 EDW77607.1 AJVK01025533 GEZM01058838 JAV71652.1 ATLV01016137 KE525057 KFB41008.1 APCN01001659 KA645018 AFP59647.1 ADMH02002089 ETN59392.1 GGFK01007075 MBW40396.1 GGFJ01003491 MBW52632.1 GGFJ01003490 MBW52631.1 GGFK01007087 MBW40408.1 AXCN02001964 GGFK01007599 MBW40920.1 GALA01000134 JAA94718.1 CH379060 EAL33796.2 CH479180 EDW29024.1 CH902624 EDV33260.1 KPU74376.1 JRES01000044 KNC34640.1 CM002910 KMY88157.1 CM000157 EDW87688.1 CH480820 EDW54285.1 CH940661 EDW71316.1 CP012523 ALC37938.1 ALC38425.1 AE014134 AY071312 AAF50965.1 AAL48934.1 GGFJ01003903 MBW53044.1 CH954177 EDV57782.1 AJWK01027315 JXJN01013398 OUUW01000004 SPP79146.1 DS231851 EDS37282.1 KQ971363 EFA07752.1 LJIG01022899 KRT78166.1 CH933807 EDW13258.1 CH916372 EDV99085.1 CCAG010010817 GL732542 EFX81645.1 GANO01000262 JAB59609.1 GDIP01054565 LRGB01001161 JAM49150.1 KZS13409.1 GDIQ01059064 JAN35673.1 LJIJ01000809 ODM94398.1 UFQS01000003 UFQT01000003 SSW96759.1 SSX17146.1 GDIP01012440 JAM91275.1 GDIP01099149 JAM04566.1 MUZQ01000072 OWK59716.1 QRBI01000104 RMC15511.1 KK755440 KFP43112.1 KL214831 KFV61958.1 AGTO01010370 AKCR02000001 PKK34258.1 KK502398 KFP23242.1 LSYS01003169 OPJ83370.1 AGCU01046922 AGCU01046923 AGCU01046924 AGCU01046925 PKK34257.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
+ More
UP000027135 UP000245037 UP000007062 UP000235965 UP000075885 UP000075881 UP000075903 UP000008820 UP000075920 UP000075902 UP000075882 UP000069940 UP000249989 UP000076407 UP000076408 UP000075900 UP000075883 UP000007798 UP000092462 UP000069272 UP000030765 UP000075840 UP000000673 UP000095301 UP000075886 UP000001819 UP000008744 UP000075884 UP000007801 UP000037069 UP000192221 UP000002282 UP000001292 UP000008792 UP000092553 UP000000803 UP000078200 UP000008711 UP000092461 UP000075880 UP000092443 UP000095300 UP000092460 UP000091820 UP000268350 UP000002320 UP000092445 UP000007266 UP000009192 UP000001070 UP000092444 UP000000305 UP000076858 UP000192223 UP000094527 UP000197619 UP000269221 UP000053875 UP000016665 UP000053872 UP000053119 UP000190648 UP000007267
UP000027135 UP000245037 UP000007062 UP000235965 UP000075885 UP000075881 UP000075903 UP000008820 UP000075920 UP000075902 UP000075882 UP000069940 UP000249989 UP000076407 UP000076408 UP000075900 UP000075883 UP000007798 UP000092462 UP000069272 UP000030765 UP000075840 UP000000673 UP000095301 UP000075886 UP000001819 UP000008744 UP000075884 UP000007801 UP000037069 UP000192221 UP000002282 UP000001292 UP000008792 UP000092553 UP000000803 UP000078200 UP000008711 UP000092461 UP000075880 UP000092443 UP000095300 UP000092460 UP000091820 UP000268350 UP000002320 UP000092445 UP000007266 UP000009192 UP000001070 UP000092444 UP000000305 UP000076858 UP000192223 UP000094527 UP000197619 UP000269221 UP000053875 UP000016665 UP000053872 UP000053119 UP000190648 UP000007267
Interpro
IPR017406
Ser/Thr_kinase_Rio3
+ More
IPR000687 RIO_kinase
IPR018935 RIO_kinase_CS
IPR011009 Kinase-like_dom_sf
IPR008266 Tyr_kinase_AS
IPR042121 MutL_C_regsub
IPR014790 MutL_C
IPR042120 MutL_C_dimsub
IPR037198 MutL_C_sf
IPR013083 Znf_RING/FYVE/PHD
IPR024610 ING_N_histone_binding
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR008831 Mediator_Med31
IPR038089 Med31_sf
IPR000719 Prot_kinase_dom
IPR000687 RIO_kinase
IPR018935 RIO_kinase_CS
IPR011009 Kinase-like_dom_sf
IPR008266 Tyr_kinase_AS
IPR042121 MutL_C_regsub
IPR014790 MutL_C
IPR042120 MutL_C_dimsub
IPR037198 MutL_C_sf
IPR013083 Znf_RING/FYVE/PHD
IPR024610 ING_N_histone_binding
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR008831 Mediator_Med31
IPR038089 Med31_sf
IPR000719 Prot_kinase_dom
ProteinModelPortal
H9JD16
A0A194R4W0
A0A2W1BLP5
A0A194QAN4
A0A2A4J0W2
A0A2H1WAA3
+ More
A0A1E1WK44 A0A0L7LQ88 S4PMA0 A0A212FK40 A0A0K8TM27 A0A1L8DLT7 A0A034V663 A0A067QUR6 A0A0K8UMG9 A0A2P8XG71 Q7Q135 A0A2J7Q9X8 A0A182P7Z4 A0A182K953 W8C3V4 A0A182UV86 Q16VU9 A0A182W8R1 A0A1Q3G358 A0A1S4FMF7 A0A182TT67 A0A182KZ44 A0A182GLE5 A0A182XGY5 A0A182Y699 A0A182G6J3 A0A0A1X499 A0A182RWQ7 A0A182LXC9 B4N0J4 A0A1B0D5R0 A0A1S4J7U9 A0A182F554 A0A1Y1LDC5 A0A084VSR4 A0A182HR90 T1PAS4 W5J4J9 A0A2M4AHY0 A0A1I8ML05 A0A2M4BHW8 A0A2M4BI49 A0A2M4AIH5 A0A182Q2S1 A0A2M4AJF5 T1E355 Q29M98 B4G9J4 A0A182NP21 B3MVJ5 A0A0L0CQJ0 A0A0J9QVS9 A0A1W4UIE4 B4NZ24 B4I178 B4MDQ5 A0A0M4E7S5 Q9VR42 A0A1A9UR86 A0A2M4BJ61 B3N4B2 A0A1B0CT99 A0A182IVQ1 A0A1A9YPR6 A0A1I8P092 A0A1B0BFF2 A0A1A9WUJ8 A0A3B0K0K4 B0W708 A0A1A9Z542 D6WVA9 A0A0T6AT21 B4KGQ8 B4JQA5 A0A1B0FN97 E9GFI5 U5EX34 A0A0P5YUC9 A0A1W4X9G6 A0A0P6ESE4 A0A1D2MN83 A0A336JWB3 A0A0P6BNV1 A0A0P5VJF9 A0A218V1A6 A0A3M0KS59 A0A091KSY3 A0A093FYQ7 U3JUF5 A0A2I0MX49 A0A091JRL0 A0A1V4KFX1 K7FFS5 A0A2I0MX50
A0A1E1WK44 A0A0L7LQ88 S4PMA0 A0A212FK40 A0A0K8TM27 A0A1L8DLT7 A0A034V663 A0A067QUR6 A0A0K8UMG9 A0A2P8XG71 Q7Q135 A0A2J7Q9X8 A0A182P7Z4 A0A182K953 W8C3V4 A0A182UV86 Q16VU9 A0A182W8R1 A0A1Q3G358 A0A1S4FMF7 A0A182TT67 A0A182KZ44 A0A182GLE5 A0A182XGY5 A0A182Y699 A0A182G6J3 A0A0A1X499 A0A182RWQ7 A0A182LXC9 B4N0J4 A0A1B0D5R0 A0A1S4J7U9 A0A182F554 A0A1Y1LDC5 A0A084VSR4 A0A182HR90 T1PAS4 W5J4J9 A0A2M4AHY0 A0A1I8ML05 A0A2M4BHW8 A0A2M4BI49 A0A2M4AIH5 A0A182Q2S1 A0A2M4AJF5 T1E355 Q29M98 B4G9J4 A0A182NP21 B3MVJ5 A0A0L0CQJ0 A0A0J9QVS9 A0A1W4UIE4 B4NZ24 B4I178 B4MDQ5 A0A0M4E7S5 Q9VR42 A0A1A9UR86 A0A2M4BJ61 B3N4B2 A0A1B0CT99 A0A182IVQ1 A0A1A9YPR6 A0A1I8P092 A0A1B0BFF2 A0A1A9WUJ8 A0A3B0K0K4 B0W708 A0A1A9Z542 D6WVA9 A0A0T6AT21 B4KGQ8 B4JQA5 A0A1B0FN97 E9GFI5 U5EX34 A0A0P5YUC9 A0A1W4X9G6 A0A0P6ESE4 A0A1D2MN83 A0A336JWB3 A0A0P6BNV1 A0A0P5VJF9 A0A218V1A6 A0A3M0KS59 A0A091KSY3 A0A093FYQ7 U3JUF5 A0A2I0MX49 A0A091JRL0 A0A1V4KFX1 K7FFS5 A0A2I0MX50
PDB
6G5I
E-value=4.34152e-58,
Score=571
Ontologies
GO
GO:0046872
GO:0004674
GO:0005524
GO:0005829
GO:0030490
GO:0030688
GO:0006298
GO:0004672
GO:0005634
GO:0005737
GO:0006468
GO:0006325
GO:0003712
GO:0006355
GO:0016592
GO:1990786
GO:0089720
GO:0071359
GO:0045089
GO:0032728
GO:0039534
GO:0098586
GO:0043124
GO:0032463
GO:0006412
GO:0016020
GO:0004702
GO:0007178
GO:0051015
GO:0003779
GO:0016021
Topology
Subcellular location
Nucleus
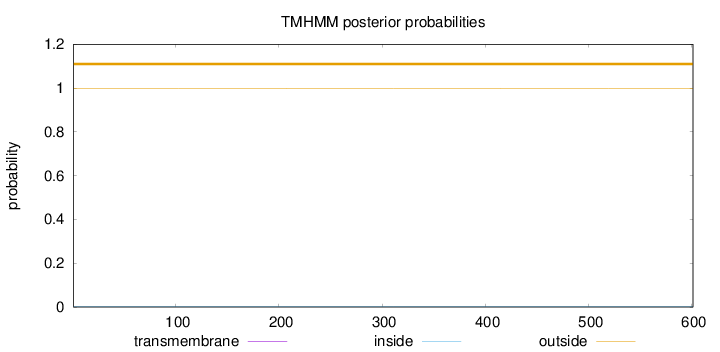
Length:
601
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00108
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00220
outside
1 - 601
Population Genetic Test Statistics
Pi
243.136268
Theta
189.890615
Tajima's D
0.453847
CLR
0.003402
CSRT
0.503774811259437
Interpretation
Uncertain
