Gene
KWMTBOMO01061 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007409
Annotation
3-hydroxyacyl-CoA_dehydrogenase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.645
Sequence
CDS
ATGATGGAGTGGGGAAAGTCGGTCGGCAAGACCTGTATCACTTGCAAGGACACCCCTGGTTTCGTTGTGAATAGACTGTTGGTGCCTTACATATGTGAAGCCATTAGGTTGTACGAAAGAGGAGATGCATCAGCTCGAGACATTGATATAGCAATGAAGCTCGGCGCTGGCTACCCCATGGGTCCACTAGAACTGGCCGACTATGTTGGCTTAGACACAAACAAGTTTATCTTGGATGGCTGGCACAAGAAGTTTCCGGATCAGCAACTCTTCAAACCTATACCCTTGCTAGAAAAGCTTGTTGCTGAAGGAAAATTAGGTGTGAAGACTGGGGAAGGTTTCTACAAATACGAAAAGAAATAA
Protein
MMEWGKSVGKTCITCKDTPGFVVNRLLVPYICEAIRLYERGDASARDIDIAMKLGAGYPMGPLELADYVGLDTNKFILDGWHKKFPDQQLFKPIPLLEKLVAEGKLGVKTGEGFYKYEKK
Summary
Uniprot
E9NYX5
H9JD12
A0A0L7LKH8
A0A2W1BJH9
A0A2H1VFH8
A0A194R582
+ More
A0A194QAC8 A0A068FKF7 I4DL36 A0A2A4J079 A0A212EUW0 S4P8W6 A0A0P6DWD7 A0A2P8XUD8 A0A0P6HK22 A0A0N7ZUY0 A0A0K8TLJ0 A0A0P5FNK2 A0A0P5SW38 A0A0P5CQ34 A0A0P5B598 A0A0P5KQY6 A0A0P5XWH8 A0A2J7Q956 U5EVM4 A0A164Q3S9 A0A0N8BD86 A0A1W4WRA7 A0A0P5FVV3 A0A0P5AD81 A0A0P5P3I4 A0A2J7Q954 A0A2J7Q952 A0A0P5HK78 A0A0N8DJA4 A0A1L8DYL4 A0A0N7ZZ61 A0A0P6DPQ1 A0A0P5ASC3 A0A1S4EJ38 A0A293LY25 A0A1B0C9M7 A0A0P6DMR6 A0A0T6AXY5 A0A1L8DYI5 A0A0P5MNB5 B0WI18 A0A1Q3FBD9 E9H317 R7UQ14 B8RJ37 A0A067R4R3 A0A182GVS0 Q17H10 A0A1E1XAL4 G3MNP3 A0A2R5L8S1 A0A131XLW9 A0A0C9S2F2 D6WWC6 R7URQ3 A0A224YGA0 A0A131YRV4 A0A023FJZ4 A0A1I8BPT8 A0A1B6FJH3 A0A1V9X114 L7M870 A0A2P6KS96 A0A2M4AEN2 A0A2M4AH39 V5GTA0 A0A131Y3K2 A0A1B6L7V5 A0A2M3Z628 A0A2H8TP71 A0A1B6GJP0 A0A2M4BV37 A0A212EV03 A0A1S3HUR3 A0A194QAI6 A0A2M4BVN9 W4XV06 T1DK38 A0A1X7VV03 A0A0T6AWW0 A0A2S2R4W1 A0A182M0W0 G0MGW4 A8XP12 A0A2G5SLD9 C1BQ93 A0A1S3H0U7 E0VKS4 B7QIS3 A0A0N7ZCX9 E2J7F6 W5JD80 A0A182SYZ0 A0A261CLY9
A0A194QAC8 A0A068FKF7 I4DL36 A0A2A4J079 A0A212EUW0 S4P8W6 A0A0P6DWD7 A0A2P8XUD8 A0A0P6HK22 A0A0N7ZUY0 A0A0K8TLJ0 A0A0P5FNK2 A0A0P5SW38 A0A0P5CQ34 A0A0P5B598 A0A0P5KQY6 A0A0P5XWH8 A0A2J7Q956 U5EVM4 A0A164Q3S9 A0A0N8BD86 A0A1W4WRA7 A0A0P5FVV3 A0A0P5AD81 A0A0P5P3I4 A0A2J7Q954 A0A2J7Q952 A0A0P5HK78 A0A0N8DJA4 A0A1L8DYL4 A0A0N7ZZ61 A0A0P6DPQ1 A0A0P5ASC3 A0A1S4EJ38 A0A293LY25 A0A1B0C9M7 A0A0P6DMR6 A0A0T6AXY5 A0A1L8DYI5 A0A0P5MNB5 B0WI18 A0A1Q3FBD9 E9H317 R7UQ14 B8RJ37 A0A067R4R3 A0A182GVS0 Q17H10 A0A1E1XAL4 G3MNP3 A0A2R5L8S1 A0A131XLW9 A0A0C9S2F2 D6WWC6 R7URQ3 A0A224YGA0 A0A131YRV4 A0A023FJZ4 A0A1I8BPT8 A0A1B6FJH3 A0A1V9X114 L7M870 A0A2P6KS96 A0A2M4AEN2 A0A2M4AH39 V5GTA0 A0A131Y3K2 A0A1B6L7V5 A0A2M3Z628 A0A2H8TP71 A0A1B6GJP0 A0A2M4BV37 A0A212EV03 A0A1S3HUR3 A0A194QAI6 A0A2M4BVN9 W4XV06 T1DK38 A0A1X7VV03 A0A0T6AWW0 A0A2S2R4W1 A0A182M0W0 G0MGW4 A8XP12 A0A2G5SLD9 C1BQ93 A0A1S3H0U7 E0VKS4 B7QIS3 A0A0N7ZCX9 E2J7F6 W5JD80 A0A182SYZ0 A0A261CLY9
Pubmed
EMBL
HQ833817
ADW77640.1
BABH01027674
JTDY01000766
KOB75952.1
KZ150083
+ More
PZC73824.1 ODYU01002291 SOQ39585.1 KQ460685 KPJ12837.1 KQ459232 KPJ02488.1 KJ622104 AID66694.1 AK402004 BAM18626.1 NWSH01004652 PCG64812.1 AGBW02012284 OWR45288.1 GAIX01004004 JAA88556.1 GDIQ01075153 JAN19584.1 PYGN01001331 PSN35623.1 GDIQ01017985 JAN76752.1 GDIP01210097 JAJ13305.1 GDAI01002582 JAI15021.1 GDIQ01252151 JAJ99573.1 GDIP01137504 JAL66210.1 GDIP01171103 JAJ52299.1 GDIP01189446 JAJ33956.1 GDIQ01187769 JAK63956.1 GDIP01078957 JAM24758.1 NEVH01016943 PNF25118.1 GANO01001813 JAB58058.1 LRGB01002485 KZS07407.1 GDIQ01189171 JAK62554.1 GDIQ01250753 JAK00972.1 GDIP01200749 JAJ22653.1 GDIQ01140592 JAL11134.1 PNF25116.1 PNF25115.1 GDIQ01249196 JAK02529.1 GDIP01028215 JAM75500.1 GFDF01002567 JAV11517.1 GDIP01198249 JAJ25153.1 GDIQ01075154 JAN19583.1 GDIP01195152 JAJ28250.1 GFWV01008843 MAA33572.1 AJWK01002493 AJWK01002494 GDIQ01075152 JAN19585.1 LJIG01022602 KRT79701.1 GFDF01002566 JAV11518.1 GDIQ01154231 JAK97494.1 DS231941 EDS28100.1 GFDL01010175 JAV24870.1 GL732587 EFX73881.1 AMQN01021812 KB299170 ELU08290.1 EZ000462 ACJ64336.1 KK852699 KDR18231.1 JXUM01091695 JXUM01091696 KQ563935 KXJ73045.1 CH477254 EAT45913.1 GFAC01002901 JAT96287.1 JO843494 AEO35111.1 GGLE01001744 MBY05870.1 GEFH01001895 JAP66686.1 GBZX01001561 JAG91179.1 KQ971361 EFA08151.1 AMQN01007360 AMQN01007361 KB300512 ELU06582.1 GFPF01005550 MAA16696.1 GEDV01006574 JAP81983.1 GBBK01003052 JAC21430.1 GECZ01019429 JAS50340.1 MNPL01029600 OQR67194.1 GACK01005671 JAA59363.1 MWRG01006094 PRD29186.1 GGFK01005914 MBW39235.1 GGFK01006785 MBW40106.1 GALX01003589 JAB64877.1 GEFM01003315 JAP72481.1 GEBQ01020252 JAT19725.1 GGFM01003193 MBW23944.1 GFXV01003233 MBW15038.1 GECZ01028505 GECZ01027315 GECZ01007115 JAS41264.1 JAS42454.1 JAS62654.1 GGFJ01007670 MBW56811.1 OWR45287.1 KPJ02487.1 GGFJ01007667 MBW56808.1 AAGJ04005685 GAMD01001141 JAB00450.1 KRT79696.1 GGMS01015189 MBY84392.1 AXCM01004407 GL379794 EGT58024.1 HE600961 CAP34252.1 PDUG01000006 PIC15729.1 BT076772 ACO11196.1 DS235250 EEB13980.1 ABJB010622494 ABJB010803024 ABJB011090822 DS947584 EEC18745.1 GDRN01057402 JAI65696.1 HP429374 ADN29874.1 ADMH02001498 ETN62302.1 NIPN01000003 OZG23055.1
PZC73824.1 ODYU01002291 SOQ39585.1 KQ460685 KPJ12837.1 KQ459232 KPJ02488.1 KJ622104 AID66694.1 AK402004 BAM18626.1 NWSH01004652 PCG64812.1 AGBW02012284 OWR45288.1 GAIX01004004 JAA88556.1 GDIQ01075153 JAN19584.1 PYGN01001331 PSN35623.1 GDIQ01017985 JAN76752.1 GDIP01210097 JAJ13305.1 GDAI01002582 JAI15021.1 GDIQ01252151 JAJ99573.1 GDIP01137504 JAL66210.1 GDIP01171103 JAJ52299.1 GDIP01189446 JAJ33956.1 GDIQ01187769 JAK63956.1 GDIP01078957 JAM24758.1 NEVH01016943 PNF25118.1 GANO01001813 JAB58058.1 LRGB01002485 KZS07407.1 GDIQ01189171 JAK62554.1 GDIQ01250753 JAK00972.1 GDIP01200749 JAJ22653.1 GDIQ01140592 JAL11134.1 PNF25116.1 PNF25115.1 GDIQ01249196 JAK02529.1 GDIP01028215 JAM75500.1 GFDF01002567 JAV11517.1 GDIP01198249 JAJ25153.1 GDIQ01075154 JAN19583.1 GDIP01195152 JAJ28250.1 GFWV01008843 MAA33572.1 AJWK01002493 AJWK01002494 GDIQ01075152 JAN19585.1 LJIG01022602 KRT79701.1 GFDF01002566 JAV11518.1 GDIQ01154231 JAK97494.1 DS231941 EDS28100.1 GFDL01010175 JAV24870.1 GL732587 EFX73881.1 AMQN01021812 KB299170 ELU08290.1 EZ000462 ACJ64336.1 KK852699 KDR18231.1 JXUM01091695 JXUM01091696 KQ563935 KXJ73045.1 CH477254 EAT45913.1 GFAC01002901 JAT96287.1 JO843494 AEO35111.1 GGLE01001744 MBY05870.1 GEFH01001895 JAP66686.1 GBZX01001561 JAG91179.1 KQ971361 EFA08151.1 AMQN01007360 AMQN01007361 KB300512 ELU06582.1 GFPF01005550 MAA16696.1 GEDV01006574 JAP81983.1 GBBK01003052 JAC21430.1 GECZ01019429 JAS50340.1 MNPL01029600 OQR67194.1 GACK01005671 JAA59363.1 MWRG01006094 PRD29186.1 GGFK01005914 MBW39235.1 GGFK01006785 MBW40106.1 GALX01003589 JAB64877.1 GEFM01003315 JAP72481.1 GEBQ01020252 JAT19725.1 GGFM01003193 MBW23944.1 GFXV01003233 MBW15038.1 GECZ01028505 GECZ01027315 GECZ01007115 JAS41264.1 JAS42454.1 JAS62654.1 GGFJ01007670 MBW56811.1 OWR45287.1 KPJ02487.1 GGFJ01007667 MBW56808.1 AAGJ04005685 GAMD01001141 JAB00450.1 KRT79696.1 GGMS01015189 MBY84392.1 AXCM01004407 GL379794 EGT58024.1 HE600961 CAP34252.1 PDUG01000006 PIC15729.1 BT076772 ACO11196.1 DS235250 EEB13980.1 ABJB010622494 ABJB010803024 ABJB011090822 DS947584 EEC18745.1 GDRN01057402 JAI65696.1 HP429374 ADN29874.1 ADMH02001498 ETN62302.1 NIPN01000003 OZG23055.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000245037 UP000235965 UP000076858 UP000192223 UP000079169 UP000092461 UP000002320 UP000000305 UP000014760 UP000027135 UP000069940 UP000249989 UP000008820 UP000007266 UP000095281 UP000192247 UP000085678 UP000007110 UP000007879 UP000075883 UP000008068 UP000008549 UP000230233 UP000009046 UP000001555 UP000000673 UP000075901 UP000216463
UP000245037 UP000235965 UP000076858 UP000192223 UP000079169 UP000092461 UP000002320 UP000000305 UP000014760 UP000027135 UP000069940 UP000249989 UP000008820 UP000007266 UP000095281 UP000192247 UP000085678 UP000007110 UP000007879 UP000075883 UP000008068 UP000008549 UP000230233 UP000009046 UP000001555 UP000000673 UP000075901 UP000216463
Interpro
Gene 3D
ProteinModelPortal
E9NYX5
H9JD12
A0A0L7LKH8
A0A2W1BJH9
A0A2H1VFH8
A0A194R582
+ More
A0A194QAC8 A0A068FKF7 I4DL36 A0A2A4J079 A0A212EUW0 S4P8W6 A0A0P6DWD7 A0A2P8XUD8 A0A0P6HK22 A0A0N7ZUY0 A0A0K8TLJ0 A0A0P5FNK2 A0A0P5SW38 A0A0P5CQ34 A0A0P5B598 A0A0P5KQY6 A0A0P5XWH8 A0A2J7Q956 U5EVM4 A0A164Q3S9 A0A0N8BD86 A0A1W4WRA7 A0A0P5FVV3 A0A0P5AD81 A0A0P5P3I4 A0A2J7Q954 A0A2J7Q952 A0A0P5HK78 A0A0N8DJA4 A0A1L8DYL4 A0A0N7ZZ61 A0A0P6DPQ1 A0A0P5ASC3 A0A1S4EJ38 A0A293LY25 A0A1B0C9M7 A0A0P6DMR6 A0A0T6AXY5 A0A1L8DYI5 A0A0P5MNB5 B0WI18 A0A1Q3FBD9 E9H317 R7UQ14 B8RJ37 A0A067R4R3 A0A182GVS0 Q17H10 A0A1E1XAL4 G3MNP3 A0A2R5L8S1 A0A131XLW9 A0A0C9S2F2 D6WWC6 R7URQ3 A0A224YGA0 A0A131YRV4 A0A023FJZ4 A0A1I8BPT8 A0A1B6FJH3 A0A1V9X114 L7M870 A0A2P6KS96 A0A2M4AEN2 A0A2M4AH39 V5GTA0 A0A131Y3K2 A0A1B6L7V5 A0A2M3Z628 A0A2H8TP71 A0A1B6GJP0 A0A2M4BV37 A0A212EV03 A0A1S3HUR3 A0A194QAI6 A0A2M4BVN9 W4XV06 T1DK38 A0A1X7VV03 A0A0T6AWW0 A0A2S2R4W1 A0A182M0W0 G0MGW4 A8XP12 A0A2G5SLD9 C1BQ93 A0A1S3H0U7 E0VKS4 B7QIS3 A0A0N7ZCX9 E2J7F6 W5JD80 A0A182SYZ0 A0A261CLY9
A0A194QAC8 A0A068FKF7 I4DL36 A0A2A4J079 A0A212EUW0 S4P8W6 A0A0P6DWD7 A0A2P8XUD8 A0A0P6HK22 A0A0N7ZUY0 A0A0K8TLJ0 A0A0P5FNK2 A0A0P5SW38 A0A0P5CQ34 A0A0P5B598 A0A0P5KQY6 A0A0P5XWH8 A0A2J7Q956 U5EVM4 A0A164Q3S9 A0A0N8BD86 A0A1W4WRA7 A0A0P5FVV3 A0A0P5AD81 A0A0P5P3I4 A0A2J7Q954 A0A2J7Q952 A0A0P5HK78 A0A0N8DJA4 A0A1L8DYL4 A0A0N7ZZ61 A0A0P6DPQ1 A0A0P5ASC3 A0A1S4EJ38 A0A293LY25 A0A1B0C9M7 A0A0P6DMR6 A0A0T6AXY5 A0A1L8DYI5 A0A0P5MNB5 B0WI18 A0A1Q3FBD9 E9H317 R7UQ14 B8RJ37 A0A067R4R3 A0A182GVS0 Q17H10 A0A1E1XAL4 G3MNP3 A0A2R5L8S1 A0A131XLW9 A0A0C9S2F2 D6WWC6 R7URQ3 A0A224YGA0 A0A131YRV4 A0A023FJZ4 A0A1I8BPT8 A0A1B6FJH3 A0A1V9X114 L7M870 A0A2P6KS96 A0A2M4AEN2 A0A2M4AH39 V5GTA0 A0A131Y3K2 A0A1B6L7V5 A0A2M3Z628 A0A2H8TP71 A0A1B6GJP0 A0A2M4BV37 A0A212EV03 A0A1S3HUR3 A0A194QAI6 A0A2M4BVN9 W4XV06 T1DK38 A0A1X7VV03 A0A0T6AWW0 A0A2S2R4W1 A0A182M0W0 G0MGW4 A8XP12 A0A2G5SLD9 C1BQ93 A0A1S3H0U7 E0VKS4 B7QIS3 A0A0N7ZCX9 E2J7F6 W5JD80 A0A182SYZ0 A0A261CLY9
PDB
4J0F
E-value=2.06433e-47,
Score=470
Ontologies
PATHWAY
00062
Fatty acid elongation - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
GO
Topology
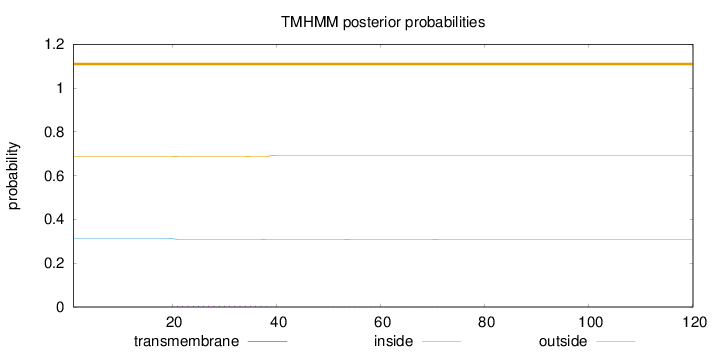
Length:
120
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10138
Exp number, first 60 AAs:
0.0972
Total prob of N-in:
0.31266
outside
1 - 120
Population Genetic Test Statistics
Pi
363.380287
Theta
204.086829
Tajima's D
2.15717
CLR
0.189661
CSRT
0.907704614769262
Interpretation
Uncertain
