Gene
KWMTBOMO01060 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007408
Annotation
3-hydroxyacyl-CoA_dehydrogenase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.427 Nuclear Reliability : 1.121
Sequence
CDS
ATGCAAAGTGCTATCAAGAATGTAACGGTTATTGGAGGTGGTCTAATGGGCTCCGGCATAGCCCAGGTATCTGCCCAAGCAGGACAGAATGTTACCCTGGTTGACGTCAGCAATGATGCTTTGGCTAAAGCCAAGAAGAGCATTGGCACAAACCTCAGCAGGGTCGCAAAGAAGATGTACAAAGATAATCCCCAAGAAGGAGAGAAATTTGTGAATGACTCACTAGGAAGGATTAATACTGCAACTGATGCAGCAGAGGCTTCCAAATCTGCTGATCTGGTTGTTGAGGCTATTGTTGAAAATATTGAAGTGAAGCATAAATTGTTTAAGCAGCTTGATGGCGTAGCTCCAAGTCACACTATTTTCGCATCAAACACGTCTTCTCTATCTATAAATGAAATTGCATCCGTCGTAAAAAGGAAAGACAAGTAG
Protein
MQSAIKNVTVIGGGLMGSGIAQVSAQAGQNVTLVDVSNDALAKAKKSIGTNLSRVAKKMYKDNPQEGEKFVNDSLGRINTATDAAEASKSADLVVEAIVENIEVKHKLFKQLDGVAPSHTIFASNTSSLSINEIASVVKRKDK
Summary
Uniprot
H9JD11
E9NYX5
A0A2H1VFH8
A0A068FKF7
A0A2W1BJH9
A0A2A4J079
+ More
A0A194QAC8 I4DL36 A0A194R582 S4P8W6 A0A0T6AXY5 A0A212EUW0 D6WWC6 A0A1Y1MXK4 A0A2S2R4W1 A0A2H8TP71 X1WJP5 C4WRK9 A0A067R4R3 R4V4D7 A0A2J7Q956 A0A2J7Q952 A0A0L7LKH8 A0A2P8XUD8 A0A2S2PDL5 A0A0T6AWW0 N6TPC9 A0A1W4WRA7 A0A087U399 V5GTA0 A0A1S3H0U7 A0A1W4WGI0 A0A2P6KS96 A0A1B6L7V5 A0A0P4YSW1 A0A0A9X4G2 H9JCW3 A0A2P8XUE6 A0A1B6HGT3 A0A0P5VHN6 A0A1B6JNA4 A0A0N8A0S4 A0A1B6J655 Q1HPZ9 R7UKI9 R4G8E3 A0A0P6FY44 A0A0N8EDM6 T1I645 A0A1B6EFT9 A0A164Q3S9 A0A0N7ZZ61 A0A0P6EYE7 A0A0N8DJA4 A0A0P6DPQ1 A0A0P5ASC3 A0A0P5P3I4 A0A1D1W5L3 A0A146LYP2 A0A1B6FJH3 A0A194QAI6 A0A1B6GJP0 A0A0P5HK78 R7URQ3 A0A0P5AVT6 A0A0K8TLJ0 A0A0P6DWD7 A0A0P5AD81 A0A0P6HK22 E9H317 Q7T0N8 R7UKY5 H3A1I3 A0A1B6G4Z2 A0A2W1BJF7 A0A1Y1L3D7 A0A2B4STZ8 E0VKS4 A0A2J8XQC5 A0A3M6TGA8 A0A2T7Q046 A0A212EV03 A0A2G8JGT9 B6RB36 A0A3P8QU67 A0A3B4FUP1 A0A3P9CW47 V4B5W7 A0A182M0W0 A0A0P5LJN5 H2PE29 Q5XGI2 A0A2J8XQM3 A0A1W4YT96 A0A2I3HVM5 H3DKZ1 I3LYG1 W5KSG6 A0A194RAE8 A0A2J8M2I1
A0A194QAC8 I4DL36 A0A194R582 S4P8W6 A0A0T6AXY5 A0A212EUW0 D6WWC6 A0A1Y1MXK4 A0A2S2R4W1 A0A2H8TP71 X1WJP5 C4WRK9 A0A067R4R3 R4V4D7 A0A2J7Q956 A0A2J7Q952 A0A0L7LKH8 A0A2P8XUD8 A0A2S2PDL5 A0A0T6AWW0 N6TPC9 A0A1W4WRA7 A0A087U399 V5GTA0 A0A1S3H0U7 A0A1W4WGI0 A0A2P6KS96 A0A1B6L7V5 A0A0P4YSW1 A0A0A9X4G2 H9JCW3 A0A2P8XUE6 A0A1B6HGT3 A0A0P5VHN6 A0A1B6JNA4 A0A0N8A0S4 A0A1B6J655 Q1HPZ9 R7UKI9 R4G8E3 A0A0P6FY44 A0A0N8EDM6 T1I645 A0A1B6EFT9 A0A164Q3S9 A0A0N7ZZ61 A0A0P6EYE7 A0A0N8DJA4 A0A0P6DPQ1 A0A0P5ASC3 A0A0P5P3I4 A0A1D1W5L3 A0A146LYP2 A0A1B6FJH3 A0A194QAI6 A0A1B6GJP0 A0A0P5HK78 R7URQ3 A0A0P5AVT6 A0A0K8TLJ0 A0A0P6DWD7 A0A0P5AD81 A0A0P6HK22 E9H317 Q7T0N8 R7UKY5 H3A1I3 A0A1B6G4Z2 A0A2W1BJF7 A0A1Y1L3D7 A0A2B4STZ8 E0VKS4 A0A2J8XQC5 A0A3M6TGA8 A0A2T7Q046 A0A212EV03 A0A2G8JGT9 B6RB36 A0A3P8QU67 A0A3B4FUP1 A0A3P9CW47 V4B5W7 A0A182M0W0 A0A0P5LJN5 H2PE29 Q5XGI2 A0A2J8XQM3 A0A1W4YT96 A0A2I3HVM5 H3DKZ1 I3LYG1 W5KSG6 A0A194RAE8 A0A2J8M2I1
Pubmed
EMBL
BABH01027674
HQ833817
ADW77640.1
ODYU01002291
SOQ39585.1
KJ622104
+ More
AID66694.1 KZ150083 PZC73824.1 NWSH01004652 PCG64812.1 KQ459232 KPJ02488.1 AK402004 BAM18626.1 KQ460685 KPJ12837.1 GAIX01004004 JAA88556.1 LJIG01022602 KRT79701.1 AGBW02012284 OWR45288.1 KQ971361 EFA08151.1 GEZM01020503 JAV89260.1 GGMS01015189 MBY84392.1 GFXV01003233 MBW15038.1 ABLF02028250 AK339814 BAH70529.1 KK852699 KDR18231.1 KC740894 AGM32718.1 NEVH01016943 PNF25118.1 PNF25115.1 JTDY01000766 KOB75952.1 PYGN01001331 PSN35623.1 GGMR01014659 MBY27278.1 KRT79696.1 APGK01056522 KB741277 KB632408 ENN71065.1 ERL95153.1 KK117947 KFM71838.1 GALX01003589 JAB64877.1 MWRG01006094 PRD29186.1 GEBQ01020252 JAT19725.1 GDIP01227643 JAI95758.1 GBHO01028717 GBHO01028715 GBRD01017621 GBRD01017620 JAG14887.1 JAG14889.1 JAG48206.1 PSN35625.1 GECU01033881 JAS73825.1 GDIP01099811 JAM03904.1 GECU01007363 JAT00344.1 GDIP01193745 JAJ29657.1 GECU01013059 JAS94647.1 DQ443253 ABF51342.1 AMQN01007361 KB300512 ELU06583.1 GAHY01001122 JAA76388.1 GDIQ01058965 JAN35772.1 GDIQ01037060 JAN57677.1 ACPB03013771 GEDC01026746 GEDC01001250 GEDC01000548 JAS10552.1 JAS36048.1 JAS36750.1 LRGB01002485 KZS07407.1 GDIP01198249 JAJ25153.1 GDIQ01056320 JAN38417.1 GDIP01028215 JAM75500.1 GDIQ01075154 JAN19583.1 GDIP01195152 JAJ28250.1 GDIQ01140592 JAL11134.1 BDGG01000016 GAV08103.1 GDHC01006873 JAQ11756.1 GECZ01019429 JAS50340.1 KPJ02487.1 GECZ01028505 GECZ01027315 GECZ01007115 JAS41264.1 JAS42454.1 JAS62654.1 GDIQ01249196 JAK02529.1 AMQN01007360 ELU06582.1 GDIP01193744 JAJ29658.1 GDAI01002582 JAI15021.1 GDIQ01075153 JAN19584.1 GDIP01200749 JAJ22653.1 GDIQ01017985 JAN76752.1 GL732587 EFX73881.1 BC056108 CM004466 AAH56108.1 OCT99841.1 AMQN01001130 AMQN01001131 AMQN01001132 KB300094 ELU07199.1 AFYH01221734 AFYH01221735 AFYH01221736 AFYH01221737 AFYH01221738 AFYH01221739 AFYH01221740 GECZ01012263 JAS57506.1 PZC73825.1 GEZM01068584 JAV66880.1 LSMT01000018 PFX32826.1 DS235250 EEB13980.1 NDHI03003363 PNJ84237.1 RCHS01003631 RMX40435.1 PZQS01000001 PVD39042.1 OWR45287.1 MRZV01002038 PIK34929.1 EF103382 ABO26640.1 KB200521 ESP01467.1 AXCM01004407 GDIQ01172669 JAK79056.1 ABGA01122417 ABGA01122418 ABGA01122419 ABGA01122420 ABGA01122421 PNJ84234.1 AAMC01090919 BC084457 CR760691 AAH84457.1 CAJ82220.1 PNJ84238.1 ADFV01176761 AGTP01110194 KPJ12836.1 NBAG03000271 PNI53726.1
AID66694.1 KZ150083 PZC73824.1 NWSH01004652 PCG64812.1 KQ459232 KPJ02488.1 AK402004 BAM18626.1 KQ460685 KPJ12837.1 GAIX01004004 JAA88556.1 LJIG01022602 KRT79701.1 AGBW02012284 OWR45288.1 KQ971361 EFA08151.1 GEZM01020503 JAV89260.1 GGMS01015189 MBY84392.1 GFXV01003233 MBW15038.1 ABLF02028250 AK339814 BAH70529.1 KK852699 KDR18231.1 KC740894 AGM32718.1 NEVH01016943 PNF25118.1 PNF25115.1 JTDY01000766 KOB75952.1 PYGN01001331 PSN35623.1 GGMR01014659 MBY27278.1 KRT79696.1 APGK01056522 KB741277 KB632408 ENN71065.1 ERL95153.1 KK117947 KFM71838.1 GALX01003589 JAB64877.1 MWRG01006094 PRD29186.1 GEBQ01020252 JAT19725.1 GDIP01227643 JAI95758.1 GBHO01028717 GBHO01028715 GBRD01017621 GBRD01017620 JAG14887.1 JAG14889.1 JAG48206.1 PSN35625.1 GECU01033881 JAS73825.1 GDIP01099811 JAM03904.1 GECU01007363 JAT00344.1 GDIP01193745 JAJ29657.1 GECU01013059 JAS94647.1 DQ443253 ABF51342.1 AMQN01007361 KB300512 ELU06583.1 GAHY01001122 JAA76388.1 GDIQ01058965 JAN35772.1 GDIQ01037060 JAN57677.1 ACPB03013771 GEDC01026746 GEDC01001250 GEDC01000548 JAS10552.1 JAS36048.1 JAS36750.1 LRGB01002485 KZS07407.1 GDIP01198249 JAJ25153.1 GDIQ01056320 JAN38417.1 GDIP01028215 JAM75500.1 GDIQ01075154 JAN19583.1 GDIP01195152 JAJ28250.1 GDIQ01140592 JAL11134.1 BDGG01000016 GAV08103.1 GDHC01006873 JAQ11756.1 GECZ01019429 JAS50340.1 KPJ02487.1 GECZ01028505 GECZ01027315 GECZ01007115 JAS41264.1 JAS42454.1 JAS62654.1 GDIQ01249196 JAK02529.1 AMQN01007360 ELU06582.1 GDIP01193744 JAJ29658.1 GDAI01002582 JAI15021.1 GDIQ01075153 JAN19584.1 GDIP01200749 JAJ22653.1 GDIQ01017985 JAN76752.1 GL732587 EFX73881.1 BC056108 CM004466 AAH56108.1 OCT99841.1 AMQN01001130 AMQN01001131 AMQN01001132 KB300094 ELU07199.1 AFYH01221734 AFYH01221735 AFYH01221736 AFYH01221737 AFYH01221738 AFYH01221739 AFYH01221740 GECZ01012263 JAS57506.1 PZC73825.1 GEZM01068584 JAV66880.1 LSMT01000018 PFX32826.1 DS235250 EEB13980.1 NDHI03003363 PNJ84237.1 RCHS01003631 RMX40435.1 PZQS01000001 PVD39042.1 OWR45287.1 MRZV01002038 PIK34929.1 EF103382 ABO26640.1 KB200521 ESP01467.1 AXCM01004407 GDIQ01172669 JAK79056.1 ABGA01122417 ABGA01122418 ABGA01122419 ABGA01122420 ABGA01122421 PNJ84234.1 AAMC01090919 BC084457 CR760691 AAH84457.1 CAJ82220.1 PNJ84238.1 ADFV01176761 AGTP01110194 KPJ12836.1 NBAG03000271 PNI53726.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000007266
+ More
UP000007819 UP000027135 UP000235965 UP000037510 UP000245037 UP000019118 UP000030742 UP000192223 UP000054359 UP000085678 UP000014760 UP000015103 UP000076858 UP000186922 UP000000305 UP000186698 UP000008672 UP000225706 UP000009046 UP000275408 UP000245119 UP000230750 UP000265100 UP000261460 UP000265160 UP000030746 UP000075883 UP000001595 UP000008143 UP000192224 UP000001073 UP000007303 UP000005215 UP000018467
UP000007819 UP000027135 UP000235965 UP000037510 UP000245037 UP000019118 UP000030742 UP000192223 UP000054359 UP000085678 UP000014760 UP000015103 UP000076858 UP000186922 UP000000305 UP000186698 UP000008672 UP000225706 UP000009046 UP000275408 UP000245119 UP000230750 UP000265100 UP000261460 UP000265160 UP000030746 UP000075883 UP000001595 UP000008143 UP000192224 UP000001073 UP000007303 UP000005215 UP000018467
Interpro
Gene 3D
ProteinModelPortal
H9JD11
E9NYX5
A0A2H1VFH8
A0A068FKF7
A0A2W1BJH9
A0A2A4J079
+ More
A0A194QAC8 I4DL36 A0A194R582 S4P8W6 A0A0T6AXY5 A0A212EUW0 D6WWC6 A0A1Y1MXK4 A0A2S2R4W1 A0A2H8TP71 X1WJP5 C4WRK9 A0A067R4R3 R4V4D7 A0A2J7Q956 A0A2J7Q952 A0A0L7LKH8 A0A2P8XUD8 A0A2S2PDL5 A0A0T6AWW0 N6TPC9 A0A1W4WRA7 A0A087U399 V5GTA0 A0A1S3H0U7 A0A1W4WGI0 A0A2P6KS96 A0A1B6L7V5 A0A0P4YSW1 A0A0A9X4G2 H9JCW3 A0A2P8XUE6 A0A1B6HGT3 A0A0P5VHN6 A0A1B6JNA4 A0A0N8A0S4 A0A1B6J655 Q1HPZ9 R7UKI9 R4G8E3 A0A0P6FY44 A0A0N8EDM6 T1I645 A0A1B6EFT9 A0A164Q3S9 A0A0N7ZZ61 A0A0P6EYE7 A0A0N8DJA4 A0A0P6DPQ1 A0A0P5ASC3 A0A0P5P3I4 A0A1D1W5L3 A0A146LYP2 A0A1B6FJH3 A0A194QAI6 A0A1B6GJP0 A0A0P5HK78 R7URQ3 A0A0P5AVT6 A0A0K8TLJ0 A0A0P6DWD7 A0A0P5AD81 A0A0P6HK22 E9H317 Q7T0N8 R7UKY5 H3A1I3 A0A1B6G4Z2 A0A2W1BJF7 A0A1Y1L3D7 A0A2B4STZ8 E0VKS4 A0A2J8XQC5 A0A3M6TGA8 A0A2T7Q046 A0A212EV03 A0A2G8JGT9 B6RB36 A0A3P8QU67 A0A3B4FUP1 A0A3P9CW47 V4B5W7 A0A182M0W0 A0A0P5LJN5 H2PE29 Q5XGI2 A0A2J8XQM3 A0A1W4YT96 A0A2I3HVM5 H3DKZ1 I3LYG1 W5KSG6 A0A194RAE8 A0A2J8M2I1
A0A194QAC8 I4DL36 A0A194R582 S4P8W6 A0A0T6AXY5 A0A212EUW0 D6WWC6 A0A1Y1MXK4 A0A2S2R4W1 A0A2H8TP71 X1WJP5 C4WRK9 A0A067R4R3 R4V4D7 A0A2J7Q956 A0A2J7Q952 A0A0L7LKH8 A0A2P8XUD8 A0A2S2PDL5 A0A0T6AWW0 N6TPC9 A0A1W4WRA7 A0A087U399 V5GTA0 A0A1S3H0U7 A0A1W4WGI0 A0A2P6KS96 A0A1B6L7V5 A0A0P4YSW1 A0A0A9X4G2 H9JCW3 A0A2P8XUE6 A0A1B6HGT3 A0A0P5VHN6 A0A1B6JNA4 A0A0N8A0S4 A0A1B6J655 Q1HPZ9 R7UKI9 R4G8E3 A0A0P6FY44 A0A0N8EDM6 T1I645 A0A1B6EFT9 A0A164Q3S9 A0A0N7ZZ61 A0A0P6EYE7 A0A0N8DJA4 A0A0P6DPQ1 A0A0P5ASC3 A0A0P5P3I4 A0A1D1W5L3 A0A146LYP2 A0A1B6FJH3 A0A194QAI6 A0A1B6GJP0 A0A0P5HK78 R7URQ3 A0A0P5AVT6 A0A0K8TLJ0 A0A0P6DWD7 A0A0P5AD81 A0A0P6HK22 E9H317 Q7T0N8 R7UKY5 H3A1I3 A0A1B6G4Z2 A0A2W1BJF7 A0A1Y1L3D7 A0A2B4STZ8 E0VKS4 A0A2J8XQC5 A0A3M6TGA8 A0A2T7Q046 A0A212EV03 A0A2G8JGT9 B6RB36 A0A3P8QU67 A0A3B4FUP1 A0A3P9CW47 V4B5W7 A0A182M0W0 A0A0P5LJN5 H2PE29 Q5XGI2 A0A2J8XQM3 A0A1W4YT96 A0A2I3HVM5 H3DKZ1 I3LYG1 W5KSG6 A0A194RAE8 A0A2J8M2I1
PDB
3RQS
E-value=5.37149e-37,
Score=380
Ontologies
GO
Topology
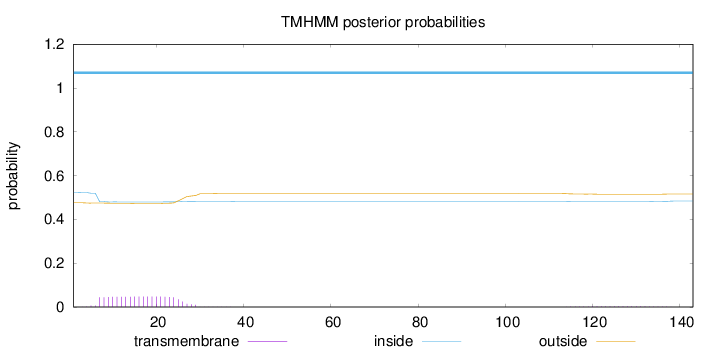
Length:
143
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.00188
Exp number, first 60 AAs:
0.94591
Total prob of N-in:
0.52390
inside
1 - 143
Population Genetic Test Statistics
Pi
279.600797
Theta
174.510425
Tajima's D
1.894438
CLR
0
CSRT
0.865056747162642
Interpretation
Uncertain
