Gene
KWMTBOMO01057 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007362
Annotation
PREDICTED:_inositol_polyphosphate_multikinase_[Papilio_xuthus]
Full name
Kinase
Location in the cell
Nuclear Reliability : 3.548
Sequence
CDS
ATGAGCGATGCGGCTGAACGACCAAAAAGGACTAAAGTGCGCCGACAGAGGTCCATCTATCAAGTGCACTATAATATCGAGAAACGCAACGACGAGCTGATGACGAAGCCCCTTGCCCTGCAGAAAGGGCTTGAAATCCAAGCCTACGATCTGCAGGTAGCCGGCCACAGAAAGACCGAAGACACCAAGTACTCAGGTTTATTGCAGTGCAGTAACGGAACAATCCTGAAACCCATAATCAAAGATTCCCAGAGGCGCGAGGTAGATTTCTATGAGCGCATGTGGGCATCGTCCGATCCTGACCTGGTGGAACTTCGAAAATGCACGCCCAAGTATTTCTACTGCAAGAAATACACTTACAATGGCTACGAACAGGAGTATATTATTCTGGAAGATCTGACGAGGAGGATGCTCGAGCCTTGCATCATGGATGTTAAAATTGGCAAAAGAACATGGGACCCGCTCGCCACAGAGGAGAAGATTAAGAACGAGCAGGCCAAGTATGCGCTGTGCAAGCAGCAGTACGGCTTCTGCATACCAGGCTTCCAGGTCTTCAAGCTGAATAGCGGGAAACTGCACAAATATAACAAGGAATACGGCAAGAAGCTCCACGGGCAAGGCGTGCGGGACGCCATAAAGAACTACCTGAACGCGATAGGCGGCGAGCTGTGTCGCGCGCTGCTGCTGCAGATCCTGTCGCAGCTGTGGCGCGTGCAGCGCTGGGCGCGCGCGCAGACCGCGCTGCGGCTGTACTCCGCCTCCGTGCTGCTGGTGTATGACGCCGCGCGCCTACGGGACTGCTGCGCCGATCACGACCTGCTCATGACCCGCAAAACATGTCGTTCCACGCCGATCACAAGGCGTCAGTCCATACCTTCATTACACAGCGGAACTTCATTCAGTGGTCAGTTGTCCAAGACAGGTCCGGTCTACAAGAAGCTGTCTTCTGTTCCCCTGTCCCCCCTCCAAAACGAGCCCCCCCATCCCCCTTTAAACTCCCCCTGGAGCGAGGCCCTCGAAAAACTCAACCACAATCACTCCTTTGAGCATAACTATGAAGATAAGTTGTCTAAAATAAAGATGAACTATCGTGCCCTTCTCGACCAATTGAGTTCGGATTCTCCCAACCCTGACCCCTGGGGTACGGTAAAAATTATCGACTTCGCTCACGCGTTCTTCAACGAGGAGGAGGAGAGGTGTGTAGATGACAATTTTAAGGACGGTATAGATTCTTTTGTAGCGATCTTCGAGGATTTCTTGAGTGAAACTGACGATCAAATTATTTGA
Protein
MSDAAERPKRTKVRRQRSIYQVHYNIEKRNDELMTKPLALQKGLEIQAYDLQVAGHRKTEDTKYSGLLQCSNGTILKPIIKDSQRREVDFYERMWASSDPDLVELRKCTPKYFYCKKYTYNGYEQEYIILEDLTRRMLEPCIMDVKIGKRTWDPLATEEKIKNEQAKYALCKQQYGFCIPGFQVFKLNSGKLHKYNKEYGKKLHGQGVRDAIKNYLNAIGGELCRALLLQILSQLWRVQRWARAQTALRLYSASVLLVYDAARLRDCCADHDLLMTRKTCRSTPITRRQSIPSLHSGTSFSGQLSKTGPVYKKLSSVPLSPLQNEPPHPPLNSPWSEALEKLNHNHSFEHNYEDKLSKIKMNYRALLDQLSSDSPNPDPWGTVKIIDFAHAFFNEEEERCVDDNFKDGIDSFVAIFEDFLSETDDQII
Summary
Similarity
Belongs to the inositol phosphokinase (IPK) family.
Uniprot
H9JCW5
A0A2H1WM27
A0A1E1WFJ8
A0A194QGA7
A0A194RAE8
A0A212EUX7
+ More
V5GJI6 D6WWC5 A0A182YC94 A0A084WIX5 B0XDK3 A0A182JMM0 A0A1L8DLV5 A0A0K8TNZ3 A0A2P8Y3D9 A0A0T6B0V2 A0A1S4F331 Q17H08 A0A0L7LI32 A0A182RAI5 A0A182N3R0 A0A2J7Q958 A0A2J7Q949 A0A1I8JVH6 E0VKS3 W5JHX6 A0A1Y9GKL4 A0A1I8JVU9 A0A182QXY6 A0A182K2S8 A0A1Y1MM02 A0A1Q3EZU1 A0A182TR19 B0WI16 A0A182WD30 A0A182P1F1 A0A182SE18 A0A1J1IX94 A0A182MAS9 A0A067RDZ2 A0A154PDX8 A0A182FTX9 A0A0M8ZRY2 A0A310SA39 A0A195C9C5 A0A2A3E8N2 A0A195FSA7 A0A158NKZ7 A0A151I2I1 N6STS8 A0A151X478 A0A0L0BL84 F4X5X2 E2AL73 E2BME3 A0A026WM76 A0A3L8DLA8 T1PC92 A0A1I8N4D4 T1EAC0
V5GJI6 D6WWC5 A0A182YC94 A0A084WIX5 B0XDK3 A0A182JMM0 A0A1L8DLV5 A0A0K8TNZ3 A0A2P8Y3D9 A0A0T6B0V2 A0A1S4F331 Q17H08 A0A0L7LI32 A0A182RAI5 A0A182N3R0 A0A2J7Q958 A0A2J7Q949 A0A1I8JVH6 E0VKS3 W5JHX6 A0A1Y9GKL4 A0A1I8JVU9 A0A182QXY6 A0A182K2S8 A0A1Y1MM02 A0A1Q3EZU1 A0A182TR19 B0WI16 A0A182WD30 A0A182P1F1 A0A182SE18 A0A1J1IX94 A0A182MAS9 A0A067RDZ2 A0A154PDX8 A0A182FTX9 A0A0M8ZRY2 A0A310SA39 A0A195C9C5 A0A2A3E8N2 A0A195FSA7 A0A158NKZ7 A0A151I2I1 N6STS8 A0A151X478 A0A0L0BL84 F4X5X2 E2AL73 E2BME3 A0A026WM76 A0A3L8DLA8 T1PC92 A0A1I8N4D4 T1EAC0
EC Number
2.7.-.-
Pubmed
EMBL
BABH01027669
ODYU01009588
SOQ54125.1
GDQN01005622
GDQN01005326
JAT85432.1
+ More
JAT85728.1 KQ459232 KPJ02486.1 KQ460685 KPJ12836.1 AGBW02012284 OWR45286.1 GALX01004207 JAB64259.1 KQ971361 EFA08152.1 ATLV01023952 KE525347 KFB50169.1 DS232763 EDS45511.1 AXCP01008854 GFDF01006645 JAV07439.1 GDAI01001737 JAI15866.1 PYGN01000975 PSN38787.1 LJIG01016373 KRT80795.1 CH477254 EAT45915.1 JTDY01000988 KOB75188.1 NEVH01016943 PNF25111.1 PNF25110.1 DS235250 EEB13979.1 ADMH02001457 ETN62510.1 APCN01003264 AXCN02001102 GEZM01027510 JAV86704.1 GFDL01014223 JAV20822.1 DS231941 EDS28098.1 CVRI01000059 CRL03185.1 AXCM01004406 AXCM01004407 KK852699 KDR18235.1 KQ434885 KZC10062.1 KQ435878 KOX69895.1 KQ762520 OAD55771.1 KQ978072 KYM97315.1 KZ288347 PBC27549.1 KQ981281 KYN43348.1 ADTU01019066 KQ976546 KYM80961.1 APGK01056522 KB741277 KB632408 ENN71064.1 ERL95152.1 KQ982557 KYQ55028.1 JRES01001701 KNC20807.1 GL888743 EGI58154.1 GL440492 EFN65828.1 GL449192 EFN83134.1 KK107152 EZA57157.1 QOIP01000007 RLU20689.1 KA645553 AFP60182.1 GAMD01000577 JAB01014.1
JAT85728.1 KQ459232 KPJ02486.1 KQ460685 KPJ12836.1 AGBW02012284 OWR45286.1 GALX01004207 JAB64259.1 KQ971361 EFA08152.1 ATLV01023952 KE525347 KFB50169.1 DS232763 EDS45511.1 AXCP01008854 GFDF01006645 JAV07439.1 GDAI01001737 JAI15866.1 PYGN01000975 PSN38787.1 LJIG01016373 KRT80795.1 CH477254 EAT45915.1 JTDY01000988 KOB75188.1 NEVH01016943 PNF25111.1 PNF25110.1 DS235250 EEB13979.1 ADMH02001457 ETN62510.1 APCN01003264 AXCN02001102 GEZM01027510 JAV86704.1 GFDL01014223 JAV20822.1 DS231941 EDS28098.1 CVRI01000059 CRL03185.1 AXCM01004406 AXCM01004407 KK852699 KDR18235.1 KQ434885 KZC10062.1 KQ435878 KOX69895.1 KQ762520 OAD55771.1 KQ978072 KYM97315.1 KZ288347 PBC27549.1 KQ981281 KYN43348.1 ADTU01019066 KQ976546 KYM80961.1 APGK01056522 KB741277 KB632408 ENN71064.1 ERL95152.1 KQ982557 KYQ55028.1 JRES01001701 KNC20807.1 GL888743 EGI58154.1 GL440492 EFN65828.1 GL449192 EFN83134.1 KK107152 EZA57157.1 QOIP01000007 RLU20689.1 KA645553 AFP60182.1 GAMD01000577 JAB01014.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000007266
UP000076408
+ More
UP000030765 UP000002320 UP000075880 UP000245037 UP000008820 UP000037510 UP000075900 UP000075884 UP000235965 UP000075903 UP000009046 UP000000673 UP000075840 UP000076407 UP000075886 UP000075881 UP000075902 UP000075920 UP000075885 UP000075901 UP000183832 UP000075883 UP000027135 UP000076502 UP000069272 UP000053105 UP000078542 UP000242457 UP000078541 UP000005205 UP000078540 UP000019118 UP000030742 UP000075809 UP000037069 UP000007755 UP000000311 UP000008237 UP000053097 UP000279307 UP000095301
UP000030765 UP000002320 UP000075880 UP000245037 UP000008820 UP000037510 UP000075900 UP000075884 UP000235965 UP000075903 UP000009046 UP000000673 UP000075840 UP000076407 UP000075886 UP000075881 UP000075902 UP000075920 UP000075885 UP000075901 UP000183832 UP000075883 UP000027135 UP000076502 UP000069272 UP000053105 UP000078542 UP000242457 UP000078541 UP000005205 UP000078540 UP000019118 UP000030742 UP000075809 UP000037069 UP000007755 UP000000311 UP000008237 UP000053097 UP000279307 UP000095301
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JCW5
A0A2H1WM27
A0A1E1WFJ8
A0A194QGA7
A0A194RAE8
A0A212EUX7
+ More
V5GJI6 D6WWC5 A0A182YC94 A0A084WIX5 B0XDK3 A0A182JMM0 A0A1L8DLV5 A0A0K8TNZ3 A0A2P8Y3D9 A0A0T6B0V2 A0A1S4F331 Q17H08 A0A0L7LI32 A0A182RAI5 A0A182N3R0 A0A2J7Q958 A0A2J7Q949 A0A1I8JVH6 E0VKS3 W5JHX6 A0A1Y9GKL4 A0A1I8JVU9 A0A182QXY6 A0A182K2S8 A0A1Y1MM02 A0A1Q3EZU1 A0A182TR19 B0WI16 A0A182WD30 A0A182P1F1 A0A182SE18 A0A1J1IX94 A0A182MAS9 A0A067RDZ2 A0A154PDX8 A0A182FTX9 A0A0M8ZRY2 A0A310SA39 A0A195C9C5 A0A2A3E8N2 A0A195FSA7 A0A158NKZ7 A0A151I2I1 N6STS8 A0A151X478 A0A0L0BL84 F4X5X2 E2AL73 E2BME3 A0A026WM76 A0A3L8DLA8 T1PC92 A0A1I8N4D4 T1EAC0
V5GJI6 D6WWC5 A0A182YC94 A0A084WIX5 B0XDK3 A0A182JMM0 A0A1L8DLV5 A0A0K8TNZ3 A0A2P8Y3D9 A0A0T6B0V2 A0A1S4F331 Q17H08 A0A0L7LI32 A0A182RAI5 A0A182N3R0 A0A2J7Q958 A0A2J7Q949 A0A1I8JVH6 E0VKS3 W5JHX6 A0A1Y9GKL4 A0A1I8JVU9 A0A182QXY6 A0A182K2S8 A0A1Y1MM02 A0A1Q3EZU1 A0A182TR19 B0WI16 A0A182WD30 A0A182P1F1 A0A182SE18 A0A1J1IX94 A0A182MAS9 A0A067RDZ2 A0A154PDX8 A0A182FTX9 A0A0M8ZRY2 A0A310SA39 A0A195C9C5 A0A2A3E8N2 A0A195FSA7 A0A158NKZ7 A0A151I2I1 N6STS8 A0A151X478 A0A0L0BL84 F4X5X2 E2AL73 E2BME3 A0A026WM76 A0A3L8DLA8 T1PC92 A0A1I8N4D4 T1EAC0
PDB
6M8E
E-value=2.44694e-25,
Score=287
Ontologies
PATHWAY
GO
PANTHER
Topology
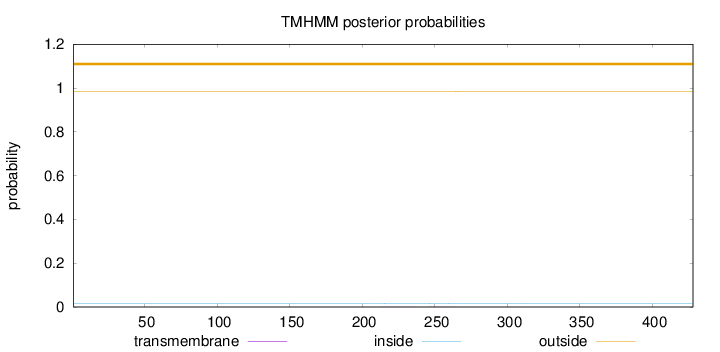
Length:
428
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0198
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01715
outside
1 - 428
Population Genetic Test Statistics
Pi
26.978285
Theta
53.021447
Tajima's D
-1.220972
CLR
0
CSRT
0.0982450877456127
Interpretation
Uncertain
