Gene
KWMTBOMO01043
Pre Gene Modal
BGIBMGA007397
Annotation
PREDICTED:_protein_charybde-like_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 1.722
Sequence
CDS
ATGGAGATCCTGCCGGTCACCAATCAATTTAATGTGGGATTCAACAGCGAAAAAGCGTGGAGCGGGCCGACATGGCGGGAAGCGCCGGCGCCGGTGCCGACGGAAGCGGCGCTGGCCCAGAGGCTCGAGAGAGAACTACGAGCGGCCAAAGGAGCCAGCGAGCTCGCCACCGCGGAGGTCTTAGTGCCGGCGGAGCTGCTCGCGAGGGCGTCGCGGCAGACCCTCGCGCTAGCGGAGGGAGAGCCCTGTGGGTCCCGGGGCGCAGCCGTCATCATCGACGTCGCGGGACGAAGACTAGCAGCCTTCAAAATAGACCCGAATACGTTGACCACGCACGAAATACATCTACACCTTGAACACGACCCCACGAACTGGACGAGTCTGTTGCCGCAATTCTTAAAAAATTTAACGCGAGGCGGCACCATCATCATCAGCCCCCAGTTCACGATAGAGAAGAAGAAATTGTTCAGGAGCCAGGCGGAGTGA
Protein
MEILPVTNQFNVGFNSEKAWSGPTWREAPAPVPTEAALAQRLERELRAAKGASELATAEVLVPAELLARASRQTLALAEGEPCGSRGAAVIIDVAGRRLAAFKIDPNTLTTHEIHLHLEHDPTNWTSLLPQFLKNLTRGGTIIISPQFTIEKKKLFRSQAE
Summary
Uniprot
S4PWM8
A0A2W1BMH8
A0A2A4JTF8
A0A2H1VGD8
A0A194R565
A0A194QAH1
+ More
A0A212FAY9 A0A0L7LKL3 H9JD00 A0A2P8ZNR6 A0A2J7QHF0 A0A088A6Z5 V9IDE3 E2AY67 A0A310SBM8 A0A067R397 A0A0J7KLU1 A0A1Y1JRU0 A0A2A3E2E8 A0A1B6CKS5 A0A2R7WRS3 A0A0M8ZNK1 A0A195FJK7 A0A158P205 E2BBQ3 A0A0P4VUK8 A0A3L8DSG2 A0A0V0G8C1 A0A0K8TN00 A0A195AVM2 A0A224XRD4 A0A069DNH5 A0A0T6BGG6 A0A0A9XKL4 A0A1B6D1N8 A0A0A9XQZ5 A0A023F4V4 D6WVR1 A0A1B6EUB9 A0A1B6MSH3 A0A0K8SDB0 A0A1W4WLE2 A0A1Y1LPS5 U5ETA3 F4W677 A0A151IKL6 A0A182FBH7 A0A1B6L203 A0A2M4BZV2 A0A2M4BZS4 A0A1B6GMC8 A0A1B0GHK0 A0A2M4BYG6 A0A1B0DC45 A0A2M4A0C6 A0A084WGN3 A0A182Q3I6 A0A2M3YX96 A0A2M3YXD4 A0A1L8DMZ6 W5J274 A0A336M831 N6TMR8 A0A0T6BCT6 A0A2S2Q4L2 A0A336KWL7 A0A182MXP6 A0A1I8MQM3 A0A1I8MQM4 A0A2S2PZ77 A0A1Y1MV34 A0A182H3R4 A0A182VL46 A0A023ELR9 A0A1A9WG84 Q7QGL3 A0A2S2N9E4 A0A1S4H717 A0A182HN67 A0A182Y628 C4WXQ7 T1E2W8 A0A1B0ANK0 A0A1S3CWT8 A0A1Q3FZ39 A0A182WEQ0 A0A1A9UPC7 A0A182RAN9 A0A1A9XCJ4 A0A182XB80 A0A1A9ZBS8 A0A1B0FG47 A0A1I8P860 A0A2H8TDK3 A0A182J3T3 B4MKN8 A0A182MHR4 B4J2V2 A0A0M5IYK9
A0A212FAY9 A0A0L7LKL3 H9JD00 A0A2P8ZNR6 A0A2J7QHF0 A0A088A6Z5 V9IDE3 E2AY67 A0A310SBM8 A0A067R397 A0A0J7KLU1 A0A1Y1JRU0 A0A2A3E2E8 A0A1B6CKS5 A0A2R7WRS3 A0A0M8ZNK1 A0A195FJK7 A0A158P205 E2BBQ3 A0A0P4VUK8 A0A3L8DSG2 A0A0V0G8C1 A0A0K8TN00 A0A195AVM2 A0A224XRD4 A0A069DNH5 A0A0T6BGG6 A0A0A9XKL4 A0A1B6D1N8 A0A0A9XQZ5 A0A023F4V4 D6WVR1 A0A1B6EUB9 A0A1B6MSH3 A0A0K8SDB0 A0A1W4WLE2 A0A1Y1LPS5 U5ETA3 F4W677 A0A151IKL6 A0A182FBH7 A0A1B6L203 A0A2M4BZV2 A0A2M4BZS4 A0A1B6GMC8 A0A1B0GHK0 A0A2M4BYG6 A0A1B0DC45 A0A2M4A0C6 A0A084WGN3 A0A182Q3I6 A0A2M3YX96 A0A2M3YXD4 A0A1L8DMZ6 W5J274 A0A336M831 N6TMR8 A0A0T6BCT6 A0A2S2Q4L2 A0A336KWL7 A0A182MXP6 A0A1I8MQM3 A0A1I8MQM4 A0A2S2PZ77 A0A1Y1MV34 A0A182H3R4 A0A182VL46 A0A023ELR9 A0A1A9WG84 Q7QGL3 A0A2S2N9E4 A0A1S4H717 A0A182HN67 A0A182Y628 C4WXQ7 T1E2W8 A0A1B0ANK0 A0A1S3CWT8 A0A1Q3FZ39 A0A182WEQ0 A0A1A9UPC7 A0A182RAN9 A0A1A9XCJ4 A0A182XB80 A0A1A9ZBS8 A0A1B0FG47 A0A1I8P860 A0A2H8TDK3 A0A182J3T3 B4MKN8 A0A182MHR4 B4J2V2 A0A0M5IYK9
Pubmed
EMBL
GAIX01006513
JAA86047.1
KZ150078
PZC73920.1
NWSH01000690
PCG74753.1
+ More
ODYU01002417 SOQ39866.1 KQ460685 KPJ12822.1 KQ459232 KPJ02472.1 AGBW02009414 OWR50883.1 JTDY01000723 KOB76093.1 BABH01027638 PYGN01000008 PSN58152.1 NEVH01013976 PNF28012.1 JR038626 JR038627 AEY58349.1 AEY58350.1 GL443762 EFN61602.1 KQ761783 OAD57050.1 KK852785 KDR16565.1 LBMM01005624 KMQ91363.1 GEZM01102330 JAV51912.1 KZ288445 PBC25654.1 GEDC01023272 JAS14026.1 KK855375 PTY22257.1 KQ435989 KOX67707.1 KQ981523 KYN40442.1 ADTU01006923 GL447151 EFN86829.1 GDKW01000214 JAI56381.1 QOIP01000005 RLU22698.1 GECL01001788 JAP04336.1 GDAI01002092 JAI15511.1 KQ976731 KYM76288.1 GFTR01002762 JAW13664.1 GBGD01003266 JAC85623.1 LJIG01000500 KRT86440.1 GBHO01022337 GDHC01009535 JAG21267.1 JAQ09094.1 GEDC01017700 JAS19598.1 GBHO01022336 GBRD01014761 GDHC01021827 JAG21268.1 JAG51065.1 JAP96801.1 GBBI01002693 JAC16019.1 KQ971358 EFA08259.1 GECZ01028217 JAS41552.1 GEBQ01001086 JAT38891.1 GBRD01014762 JAG51064.1 GEZM01055676 JAV72987.1 GANO01002870 JAB57001.1 GL887707 EGI70212.1 KQ977199 KYN05004.1 GEBQ01022302 JAT17675.1 GGFJ01009455 MBW58596.1 GGFJ01009438 MBW58579.1 GECZ01006157 JAS63612.1 AJWK01005253 GGFJ01008986 MBW58127.1 AJVK01005019 GGFK01000880 MBW34201.1 ATLV01023647 KE525345 KFB49377.1 AXCN02001050 GGFM01000148 MBW20899.1 GGFM01000160 MBW20911.1 GFDF01006258 JAV07826.1 ADMH02002136 ETN58357.1 UFQT01000531 SSX25039.1 APGK01058555 APGK01058556 KB741291 ENN70515.1 LJIG01001798 KRT85142.1 GGMS01003474 MBY72677.1 UFQS01000895 UFQT01000895 SSX07533.1 SSX27873.1 GGMS01001481 MBY70684.1 GEZM01023442 JAV88390.1 JXUM01107896 KQ565182 KXJ71246.1 GAPW01003397 JAC10201.1 AAAB01008834 EAA05686.4 GGMR01001235 MBY13854.1 APCN01001866 ABLF02033096 AK342777 BAH72677.1 GALA01000905 JAA93947.1 JXJN01000870 JXJN01000871 JXJN01000872 GFDL01002206 JAV32839.1 CCAG010003863 GFXV01000350 MBW12155.1 CH963847 EDW72744.2 AXCM01008162 CH916366 EDV97122.1 CP012525 ALC43706.1
ODYU01002417 SOQ39866.1 KQ460685 KPJ12822.1 KQ459232 KPJ02472.1 AGBW02009414 OWR50883.1 JTDY01000723 KOB76093.1 BABH01027638 PYGN01000008 PSN58152.1 NEVH01013976 PNF28012.1 JR038626 JR038627 AEY58349.1 AEY58350.1 GL443762 EFN61602.1 KQ761783 OAD57050.1 KK852785 KDR16565.1 LBMM01005624 KMQ91363.1 GEZM01102330 JAV51912.1 KZ288445 PBC25654.1 GEDC01023272 JAS14026.1 KK855375 PTY22257.1 KQ435989 KOX67707.1 KQ981523 KYN40442.1 ADTU01006923 GL447151 EFN86829.1 GDKW01000214 JAI56381.1 QOIP01000005 RLU22698.1 GECL01001788 JAP04336.1 GDAI01002092 JAI15511.1 KQ976731 KYM76288.1 GFTR01002762 JAW13664.1 GBGD01003266 JAC85623.1 LJIG01000500 KRT86440.1 GBHO01022337 GDHC01009535 JAG21267.1 JAQ09094.1 GEDC01017700 JAS19598.1 GBHO01022336 GBRD01014761 GDHC01021827 JAG21268.1 JAG51065.1 JAP96801.1 GBBI01002693 JAC16019.1 KQ971358 EFA08259.1 GECZ01028217 JAS41552.1 GEBQ01001086 JAT38891.1 GBRD01014762 JAG51064.1 GEZM01055676 JAV72987.1 GANO01002870 JAB57001.1 GL887707 EGI70212.1 KQ977199 KYN05004.1 GEBQ01022302 JAT17675.1 GGFJ01009455 MBW58596.1 GGFJ01009438 MBW58579.1 GECZ01006157 JAS63612.1 AJWK01005253 GGFJ01008986 MBW58127.1 AJVK01005019 GGFK01000880 MBW34201.1 ATLV01023647 KE525345 KFB49377.1 AXCN02001050 GGFM01000148 MBW20899.1 GGFM01000160 MBW20911.1 GFDF01006258 JAV07826.1 ADMH02002136 ETN58357.1 UFQT01000531 SSX25039.1 APGK01058555 APGK01058556 KB741291 ENN70515.1 LJIG01001798 KRT85142.1 GGMS01003474 MBY72677.1 UFQS01000895 UFQT01000895 SSX07533.1 SSX27873.1 GGMS01001481 MBY70684.1 GEZM01023442 JAV88390.1 JXUM01107896 KQ565182 KXJ71246.1 GAPW01003397 JAC10201.1 AAAB01008834 EAA05686.4 GGMR01001235 MBY13854.1 APCN01001866 ABLF02033096 AK342777 BAH72677.1 GALA01000905 JAA93947.1 JXJN01000870 JXJN01000871 JXJN01000872 GFDL01002206 JAV32839.1 CCAG010003863 GFXV01000350 MBW12155.1 CH963847 EDW72744.2 AXCM01008162 CH916366 EDV97122.1 CP012525 ALC43706.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
UP000005204
+ More
UP000245037 UP000235965 UP000005203 UP000000311 UP000027135 UP000036403 UP000242457 UP000053105 UP000078541 UP000005205 UP000008237 UP000279307 UP000078540 UP000007266 UP000192223 UP000007755 UP000078542 UP000069272 UP000092461 UP000092462 UP000030765 UP000075886 UP000000673 UP000019118 UP000075884 UP000095301 UP000069940 UP000249989 UP000075903 UP000091820 UP000007062 UP000075840 UP000076408 UP000007819 UP000092460 UP000079169 UP000075920 UP000078200 UP000075900 UP000092443 UP000076407 UP000092445 UP000092444 UP000095300 UP000075880 UP000007798 UP000075883 UP000001070 UP000092553
UP000245037 UP000235965 UP000005203 UP000000311 UP000027135 UP000036403 UP000242457 UP000053105 UP000078541 UP000005205 UP000008237 UP000279307 UP000078540 UP000007266 UP000192223 UP000007755 UP000078542 UP000069272 UP000092461 UP000092462 UP000030765 UP000075886 UP000000673 UP000019118 UP000075884 UP000095301 UP000069940 UP000249989 UP000075903 UP000091820 UP000007062 UP000075840 UP000076408 UP000007819 UP000092460 UP000079169 UP000075920 UP000078200 UP000075900 UP000092443 UP000076407 UP000092445 UP000092444 UP000095300 UP000075880 UP000007798 UP000075883 UP000001070 UP000092553
PRIDE
Pfam
PF07809 RTP801_C
Gene 3D
ProteinModelPortal
S4PWM8
A0A2W1BMH8
A0A2A4JTF8
A0A2H1VGD8
A0A194R565
A0A194QAH1
+ More
A0A212FAY9 A0A0L7LKL3 H9JD00 A0A2P8ZNR6 A0A2J7QHF0 A0A088A6Z5 V9IDE3 E2AY67 A0A310SBM8 A0A067R397 A0A0J7KLU1 A0A1Y1JRU0 A0A2A3E2E8 A0A1B6CKS5 A0A2R7WRS3 A0A0M8ZNK1 A0A195FJK7 A0A158P205 E2BBQ3 A0A0P4VUK8 A0A3L8DSG2 A0A0V0G8C1 A0A0K8TN00 A0A195AVM2 A0A224XRD4 A0A069DNH5 A0A0T6BGG6 A0A0A9XKL4 A0A1B6D1N8 A0A0A9XQZ5 A0A023F4V4 D6WVR1 A0A1B6EUB9 A0A1B6MSH3 A0A0K8SDB0 A0A1W4WLE2 A0A1Y1LPS5 U5ETA3 F4W677 A0A151IKL6 A0A182FBH7 A0A1B6L203 A0A2M4BZV2 A0A2M4BZS4 A0A1B6GMC8 A0A1B0GHK0 A0A2M4BYG6 A0A1B0DC45 A0A2M4A0C6 A0A084WGN3 A0A182Q3I6 A0A2M3YX96 A0A2M3YXD4 A0A1L8DMZ6 W5J274 A0A336M831 N6TMR8 A0A0T6BCT6 A0A2S2Q4L2 A0A336KWL7 A0A182MXP6 A0A1I8MQM3 A0A1I8MQM4 A0A2S2PZ77 A0A1Y1MV34 A0A182H3R4 A0A182VL46 A0A023ELR9 A0A1A9WG84 Q7QGL3 A0A2S2N9E4 A0A1S4H717 A0A182HN67 A0A182Y628 C4WXQ7 T1E2W8 A0A1B0ANK0 A0A1S3CWT8 A0A1Q3FZ39 A0A182WEQ0 A0A1A9UPC7 A0A182RAN9 A0A1A9XCJ4 A0A182XB80 A0A1A9ZBS8 A0A1B0FG47 A0A1I8P860 A0A2H8TDK3 A0A182J3T3 B4MKN8 A0A182MHR4 B4J2V2 A0A0M5IYK9
A0A212FAY9 A0A0L7LKL3 H9JD00 A0A2P8ZNR6 A0A2J7QHF0 A0A088A6Z5 V9IDE3 E2AY67 A0A310SBM8 A0A067R397 A0A0J7KLU1 A0A1Y1JRU0 A0A2A3E2E8 A0A1B6CKS5 A0A2R7WRS3 A0A0M8ZNK1 A0A195FJK7 A0A158P205 E2BBQ3 A0A0P4VUK8 A0A3L8DSG2 A0A0V0G8C1 A0A0K8TN00 A0A195AVM2 A0A224XRD4 A0A069DNH5 A0A0T6BGG6 A0A0A9XKL4 A0A1B6D1N8 A0A0A9XQZ5 A0A023F4V4 D6WVR1 A0A1B6EUB9 A0A1B6MSH3 A0A0K8SDB0 A0A1W4WLE2 A0A1Y1LPS5 U5ETA3 F4W677 A0A151IKL6 A0A182FBH7 A0A1B6L203 A0A2M4BZV2 A0A2M4BZS4 A0A1B6GMC8 A0A1B0GHK0 A0A2M4BYG6 A0A1B0DC45 A0A2M4A0C6 A0A084WGN3 A0A182Q3I6 A0A2M3YX96 A0A2M3YXD4 A0A1L8DMZ6 W5J274 A0A336M831 N6TMR8 A0A0T6BCT6 A0A2S2Q4L2 A0A336KWL7 A0A182MXP6 A0A1I8MQM3 A0A1I8MQM4 A0A2S2PZ77 A0A1Y1MV34 A0A182H3R4 A0A182VL46 A0A023ELR9 A0A1A9WG84 Q7QGL3 A0A2S2N9E4 A0A1S4H717 A0A182HN67 A0A182Y628 C4WXQ7 T1E2W8 A0A1B0ANK0 A0A1S3CWT8 A0A1Q3FZ39 A0A182WEQ0 A0A1A9UPC7 A0A182RAN9 A0A1A9XCJ4 A0A182XB80 A0A1A9ZBS8 A0A1B0FG47 A0A1I8P860 A0A2H8TDK3 A0A182J3T3 B4MKN8 A0A182MHR4 B4J2V2 A0A0M5IYK9
PDB
3LQ9
E-value=9.63248e-05,
Score=103
Ontologies
GO
PANTHER
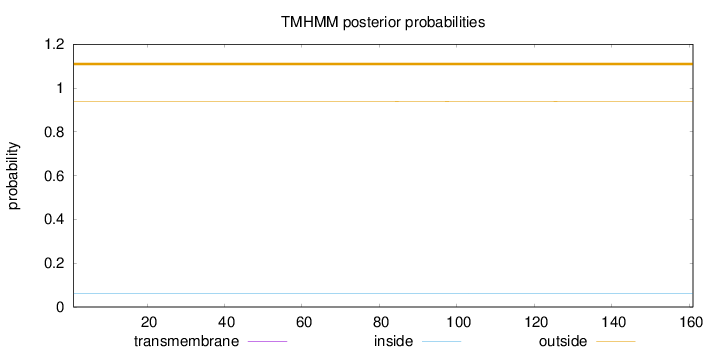
Topology
Length:
161
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00117
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.06066
outside
1 - 161
Population Genetic Test Statistics
Pi
201.163499
Theta
168.603573
Tajima's D
0.607417
CLR
0
CSRT
0.547422628868557
Interpretation
Uncertain
